
Single-Cell DNA & RNA CO-SEQUENCING Capture genotype and phenotype from the same cell — now at massive scale.

Genomic alterations influence gene expression, yet most high-throughput single-cell methods measure DNA and RNA separately. As a result, genotype–phenotype relationships are typically inferred across cells — a workaround that is insufficient to determine how genetic variation influences transcriptional heterogeneity, clonal identity, and cell state.
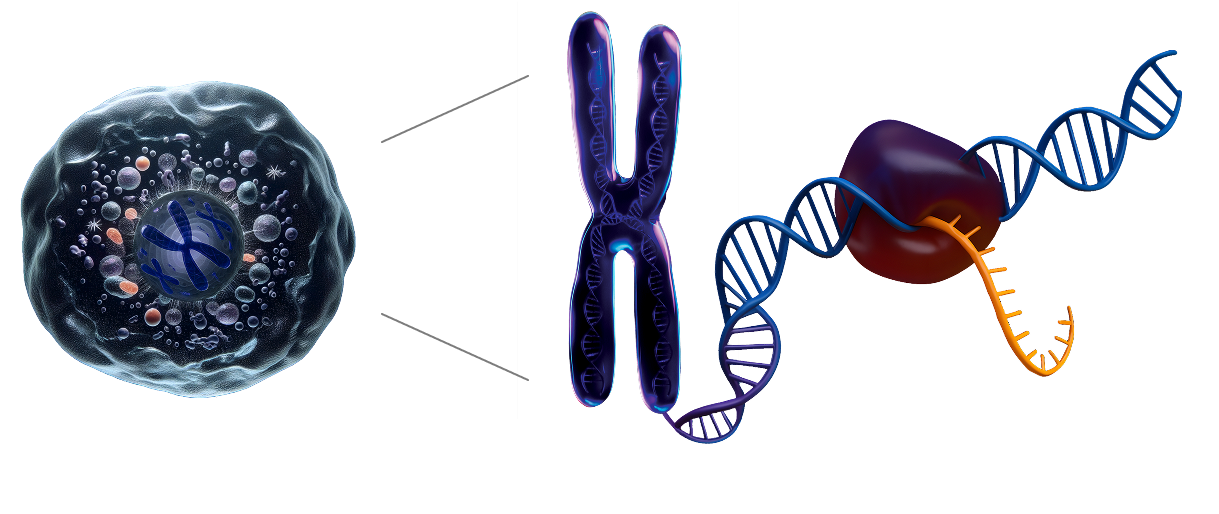
Atrandi Biosciences unveils a breakthrough platform for high-throughput single-cell DNA and RNA co-sequencing — replacing inference with direct genotype–phenotype resolution.



Joint profiling of targeted DNA variants and 3′ gene expression
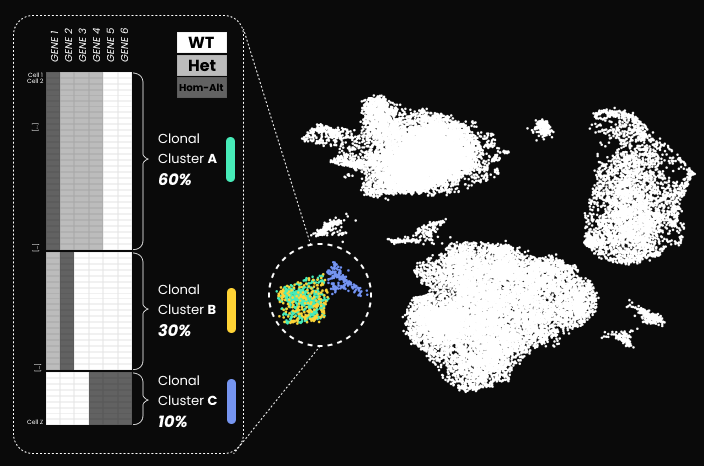
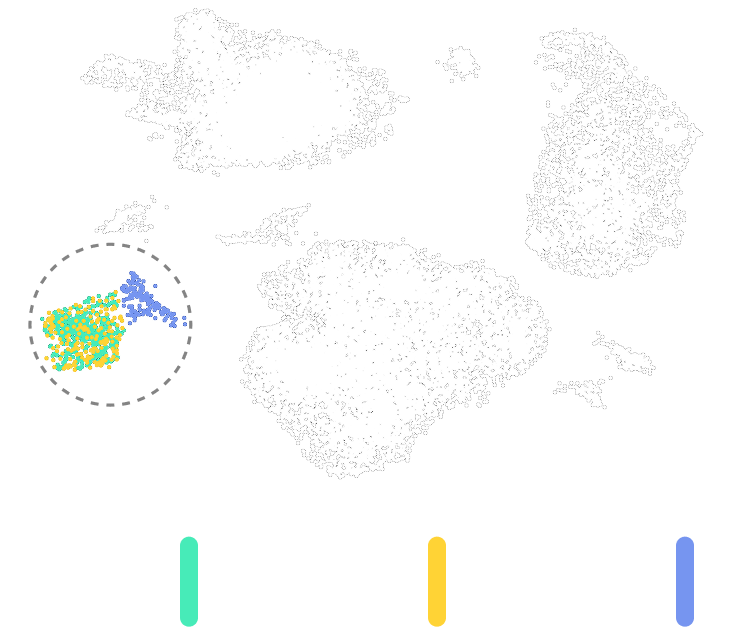
Uncover hidden clonal structure across transcriptomic clusters

Compare wild-type and variant cells to uncover genotype-specific expression programs

Reveal the distribution and frequency of engineered or naturally occurring DNA variants across transcriptionally defined cell populations
ONE FLEXIBLE SOLUTION FOR ANY NUMBER OF USER-DEFINED TARGETS
1-10 TARGETS
Confirm CRISPR edits, guide integration or trace lineage barcodes

10-100 TARGETs
Resolve clonal architecture and mutational diversity with targeted panels

100+ Targets
Profile pooled perturbation libraries or explore tumor-wide mutation landscapes
Applications
FUNCTIONAL GENOMICS

CRISPR PERTURBATION SCREENING
Confirm guide integration and on-target edits directly at the DNA level and uncover their transcriptional consequences within the same cell, supporting high-throughput functional screens without relying on guide barcodes or indirect inference.

LINEAGE TRACING
Map cellular ancestry by reading DNA-encoded lineage barcodes and connecting them to transcriptional profiles, offering high-resolution reconstruction of developmental trajectories and recording of molecular histories.
Cancer Research

TUMOR HETEROGENEITY
Characterize the genetic and phenotypic diversity of tumor cells to expose hidden cell states, regulatory plasticity, and divergent transcriptional programs that shape disease progression and therapeutic failure.
CLONAL DYNAMICS
Resolve clonal architecture and dynamics by profiling key genomic mutations and expression states from the same cell, enabling high-sensitivity tracking of selection and resistance over time.
Workflow
Features

SCale to address biological heterogeneity
Process 10,000s-1,000,000s of cells in parallel

PANEL FLEXIBILITY
Create and run custom panels yourself—starting from just one target. No restrictions.

NO COMPROMISE LYSIS
Fully disrupt cells and strip nucleosomes to access clean DNA and RNA for amplification

SEQUENCING FLEXIBILITY
Flexible sequencing of DNA and RNA - short or long reads, together or apart, your choice.
VERSATILE INPUT TYPES
Process fresh cells, fixed cells, or nuclei

Low cost for routine use
$0.01-$0.20 per cell (scale dependent)
Learn more about the SPC technology powering this workflow
PROOF OF CONCEPT
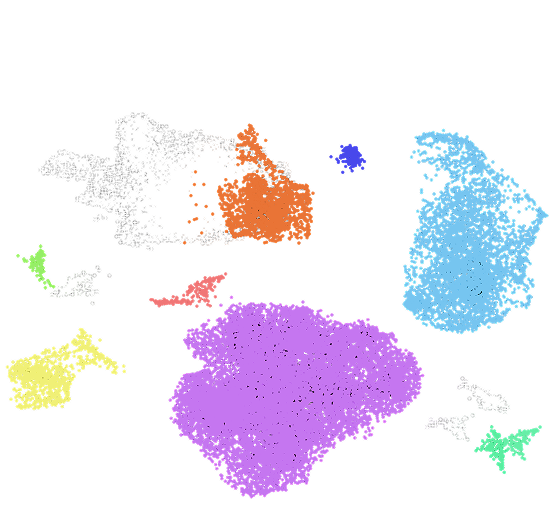
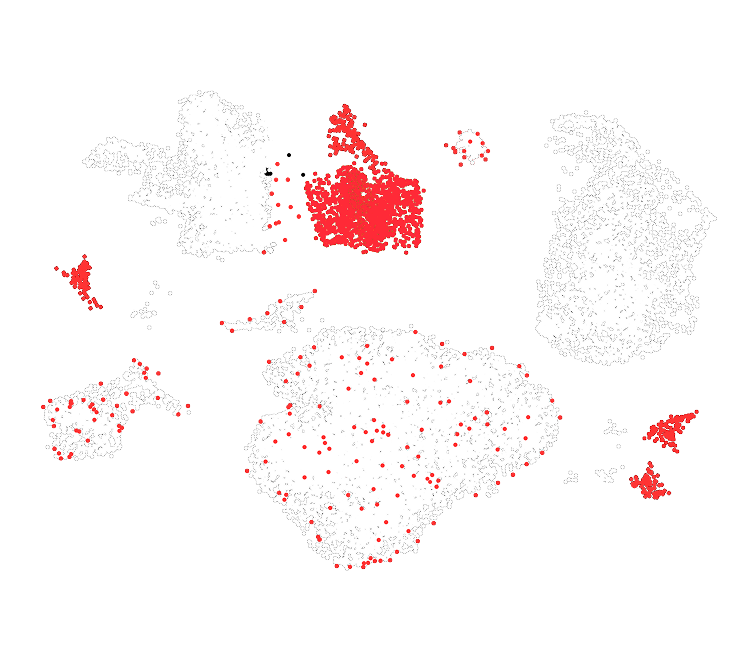
We have applied our DNA and RNA co-assay to profile healthy (PBMCs) and malignant (acute myeloid leukemia) cells, analyzing over 10,000 cells per experiment. By combining single-cell RNA-seq with targeted genomic DNA profiling using a custom PCR panel, we linked transcriptional states to precise genotypes in the same cells.

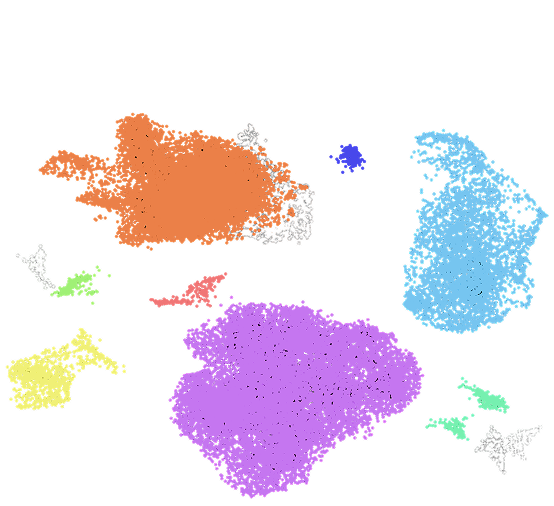
– scRNA-seq data reveals the transcriptional population structure through clustering and annotation of single cells.
– A 20-plex PCR amplicon panel enabled direct genotyping of genomic targets at single-cell resolution.
– In AML patient samples, the assay revealed rare genotypic subpopulations and their distinct transcriptional programs.