The intestinal flora consists of almost 40 trillion bacteria, not including other microbes such as fungi and archaebacteria. These microbes play a crucial role in maintaining gut health. Studies have shown an imbalance in the gut microbiome, known as gut dysbiosis, is linked to various diseases including inflammatory bowel disease, metabolic syndrome, non-alcoholic fatty liver disease, and colorectal cancer. Although our understanding of these associations has improved in recent years, further investigation is still needed to establish causation and explore the remaining 50% of intestinal bacteria that has yet to be cultured. The advancement of microfluidic technology has opened up new possibilities for studying the gut microbiome in a more precise and controlled manner. From organ-on-a-chip to single-microbe sequencing to drug delivery system development, microfluidics offers multiple ways to investigate the complexity of the gut microbiota and its impact on human health. In this review, Qi et al. delve into the diverse applications and promising potential of microfluidics in unraveling the gut microbiome and its interactions with the host.

Qi et al. first discusses the development of an intestine-on-a-chip used to simulate the physiological microenvironment of the intestine, facilitating the cultivation of gut microbiota. Previous work has developed a gut-on-a-chip system that combines epithelial and microvascular endothelial cells from intestinal biopsy samples. This platform replicates the liquid flow and peristaltic movement of the intestinal lumen. The chip consists of a flexible transparent support layer with microfluidic channels and lateral chambers separated by a porous membrane coated with an extracellular matrix. By applying cyclic suction, the central membrane stretches, inducing mechanical deformation in the monolayer of cells on it, mimicking the peristaltic motion of the intestinal lumen. Additionally, fluid flow in the chip's central microchannel can be controlled to simulate the flow in the intestinal lumen at a low rate. This chip enables the co-culture of primary human intestinal epithelium, intestinal microvascular endothelium, and intestinal bacteria, allowing for the development of four types of intestinal cells and the formation of intestinal villi and crypt structures. In one example, Kim et al. cocultured human intestinal epithelial cells (Caco-2) with Lactobacillus rhamnosus over a two-week period. These chip-based cultures demonstrated enhanced barrier function simulation, cytochrome P450 activity, and apical mucus secretion. This suggests that microfluidic-based cell culture can support cocultures with intestinal bacteria and are more physiologically relevant compared to traditional cell culture methods.
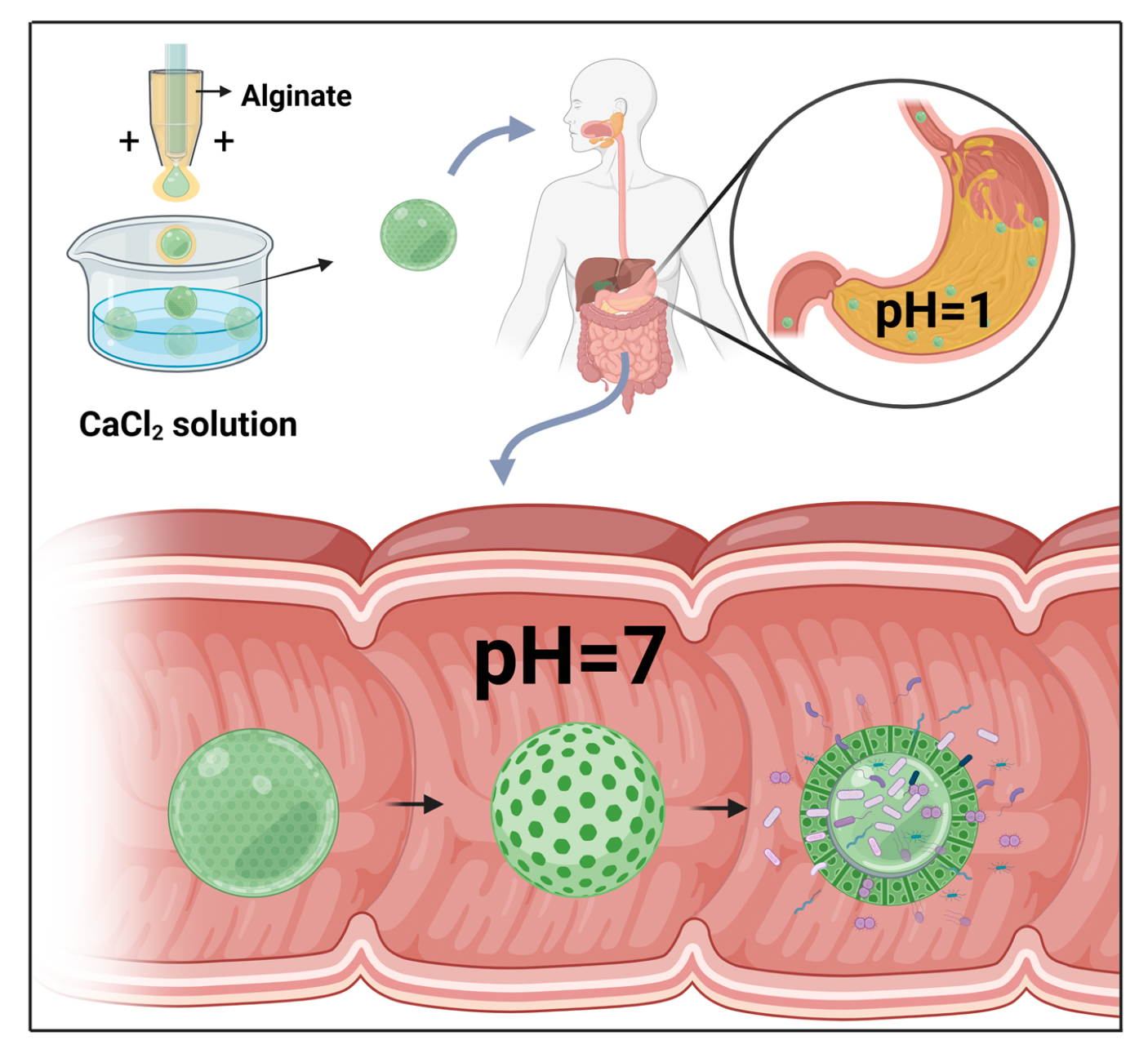
Drug delivery systems, such as nanoparticles, gel complexes, and enteric coating, are commonly used in research on intestinal microorganisms. Droplet microfluidics can synthesize specialized microcapsules with uniform size, morphology, and structure for delivering active substances like bacteriophages. These drug delivery systems enable pre-programmed and controlled release of drugs and can be used to supplement the human body with prebiotics, probiotics, and other active compounds. In one example, Zhao et al. used microfluidic electrospray chips to produce dual-core probiotic microcapsules. Alginate was used as the external shell, while microspheres containing two species of probiotic bacteria were electrosprayed as the internal phase. The microcapsules were pH-responsive, protecting the probiotics from gastric acid in the stomach and releasing them efficiently in the intestinal environment. In a mouse model of metabolic syndrome, administration of these probiotic microcapsules resulted in reduced liver fat deposits and improved intestinal barrier proteins, indicating their potential for treating the complications of metabolic syndrome. These findings indicate the immense potential of microfluidics in generating drug delivery systems suitable for the gastrointestinal tract.
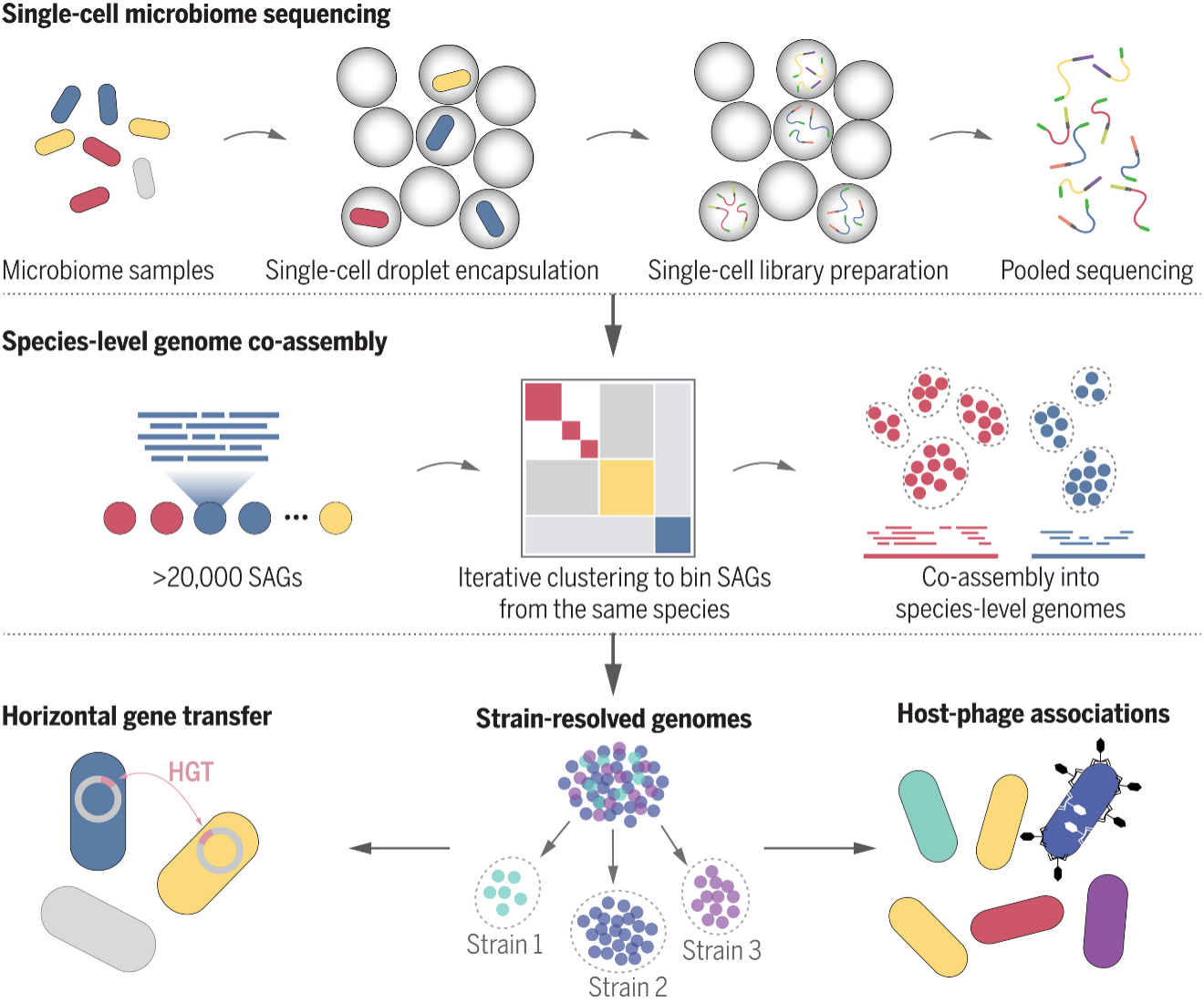
Droplet-based microfluidics has also significantly impacted microbial genomics research. 16S rRNA and metagenomic sequencing have limited bacterial phylogenetic resolution. By encapsulating microbes in droplets and performing single-cell analyses, investigators are now able to examine species- and strain-level genomes in addition to mobile elements such as plasmids and prophages. For instance, Zheng et al. used droplet microfluidics to amplify 22,000 genomes that corresponded to 76 species, many of which are difficult to culture. The authors also noted that ten of these species were of different strains. At the strain-level resolution, Zheng et al. was able to probe bacteriophage interactions and discovered that the most abundant bacteriophage crAssphage was linked to a specific strain of Bacteroides vulgatus. These interactions would have been difficult to tease apart with traditional genomic analyses. Overall, there are several advantages to implementing microfluidic technology to intestinal microbiota research. The intestine-on-a-chip system replicates the physiological conditions of the intestine, including the spatial structure of the intestinal epithelial tissue, cell interactions, and peristalsis. Compared to traditional methods, cells cultured on an intestine-on-a-chip closely resemble human duodenal epithelial cells in terms of their morphology, structure, composition, and gene expression. Similarly, there are several advantages to microfluidic drug delivery systems. These microcapsules can effectively carry living intestinal microorganisms. Microfluidic electrospray systems offer precise control over the loading and release of microorganisms into microcapsules, making them ideal delivery systems. Apart from these advantages, microfluidic platforms require a high level of technical expertise. Multiple kits and chips and droplet generators are commercially available so investigators can adopt microfluidics technology with ease.
- Qi P, Lv J, Yan X, Bai L, Zhang L. Microfluidics: Insights into Intestinal Microorganisms. Microorganisms. 2023 Apr 27;11(5):1134. doi: 10.3390/microorganisms11051134.
- Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012 Jun 21;12(12):2165-74. doi: 10.1039/c2lc40074j. Epub 2012 Mar 20. PMID: 22434367.
- Zhao C., Zhu Y., Kong B., Huang Y., Yan D., Tan H., Shang L. Dual-Core Prebiotic Microcapsule Encapsulating Probiotics for Metabolic Syndrome. ACS Appl. Mater. Interfaces. 2020;12:42586–42594. doi: 10.1021/acsami.0c13518.
- Zheng W, Zhao S, Yin Y, Zhang H, Needham DM, Evans ED, Dai CL, Lu PJ, Alm EJ, Weitz DA. High-throughput, single-microbe genomics with strain resolution, applied to a human gut microbiome. Science. 2022 Jun 3;376(6597):eabm1483. doi: 10.1126/science.abm1483. Epub 2022 Jun 3. PMID: 35653470.


