High-throughput single-cell sequencing has greatly advanced the field of genomics by allowing researchers to investigate cellular heterogeneity. However, the use of cell barcodes for identifying individual cells is limited by the cost and technical challenges associated with generating unique sets of DNA oligonucleotides for each individual cell. While this approach works well for studies with high target numbers, it becomes extremely difficult in the case of trying to detect low target numbers among thousands or millions of cells. In such cases, the use of cellular barcodes becomes costly and technically challenging. To overcome this limitation, Atrandi Biosciences has developed an alternative strategy based on the 1-read-1-cell principle, which enables sequencing of large numbers of cells in a barcoding-free fashion. This approach has the potential to reduce the cost and complexity of single-cell sequencing studies, making it more accessible and feasible to study multiple targets simultaneously.
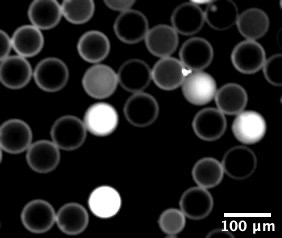
To demonstrate the feasibility of this new principle, the 1-read-1-cell paradigm was applied to recovering native B-cell receptor heavy and light chain pairs in single cells, a critical step to monoclonal antibody discovery. Monoclonal antibodies are widely used in research, diagnostics, and therapeutics. Monoclonal antibodies are produced by immortalizing a single specific B cell, which requires identifying and isolating individual B cells. First, antibody-secreting mouse hybridoma cells were sequestered in semi-permeable capsules (SPCs) based on droplet microfluidic technology. SPCs are designed to enable multiple-step reactions while still maintaining the compartmentalization of individual cells. DNA or cDNA from reverse transcriptase reactions are then amplified within each SPC. No barcoding is necessary because the 1-read-1-cell principle is applied.
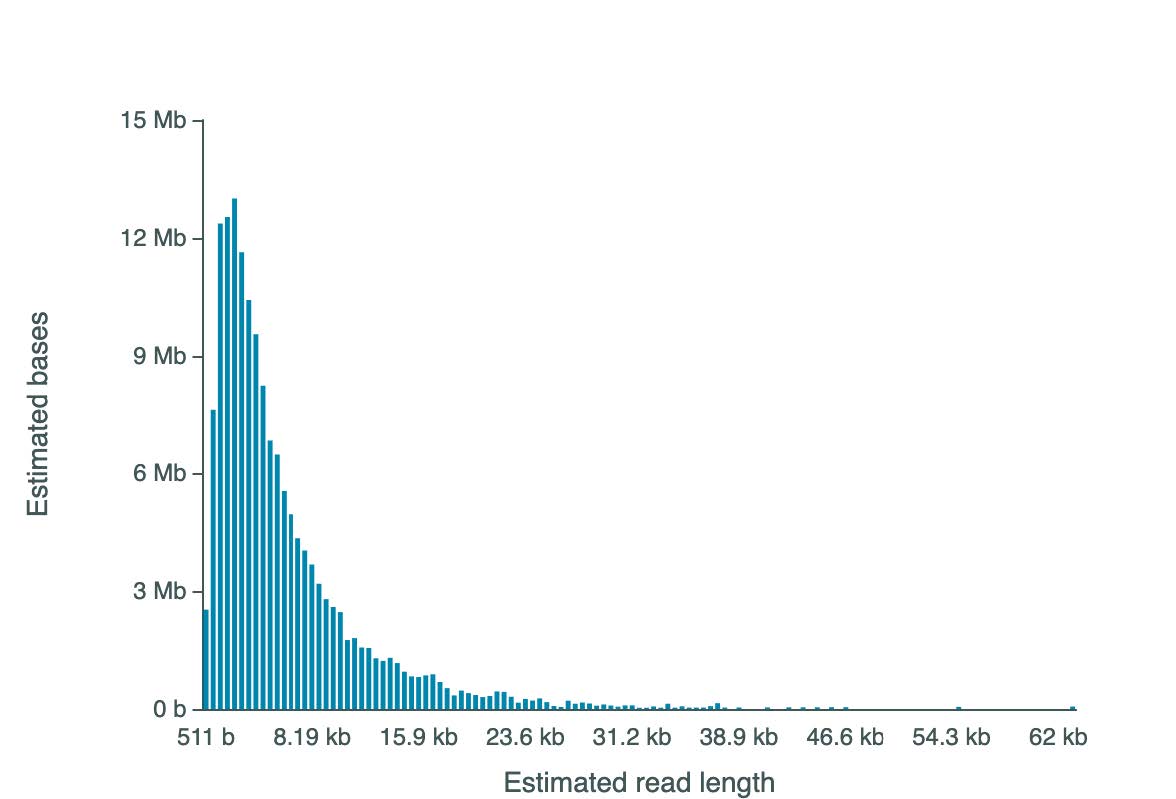
Instead of using barcodes, short DNA amplicons from the same cell were concatenated into a single fragment that then can be sequenced using long-read sequencing technology. These single-cell-derived concatemeric reads encode both heavy and light chain sequences. Results showed that the majority of concatemeric Nanopore read length ranged from 511 bases to ~24,000 bases.
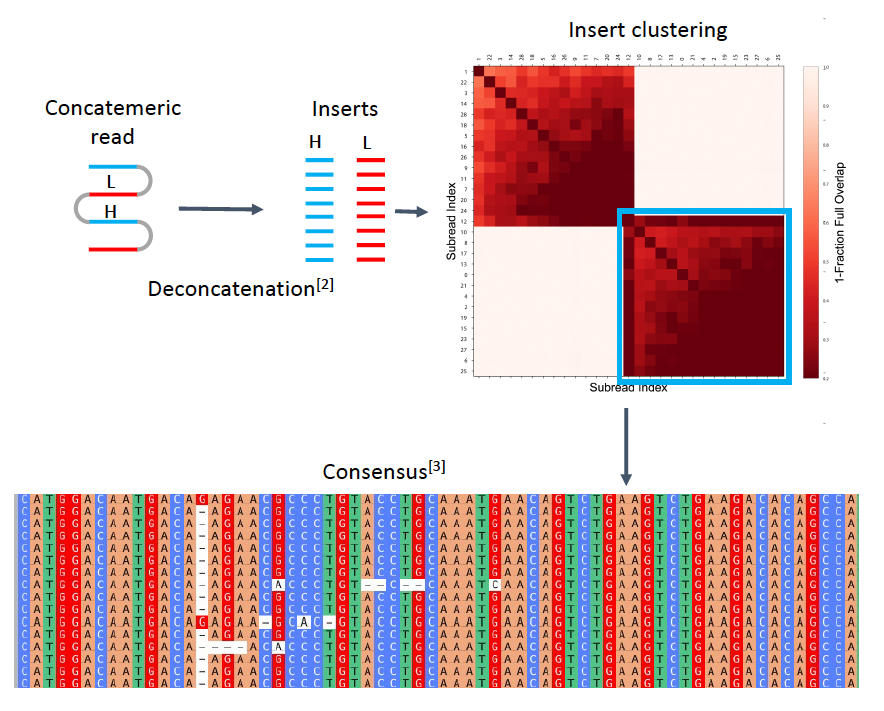
To analyze heavy and light chain inserts, concatemers were deconcatenated and clustered in silico into two groups based on sequence similarity. As the authors expected, the BCR heavy and light chain inserts clustered independently of each other. More importantly, because of insert redundancy in concatemers, the authors were able to correct sequencing errors by consensus nucleotide alignment. As a proof-of-concept, the authors successfully employed a barcoding-free approach to sequence BCR heavy and light chain pairs in individual cells, a vital stage in monoclonal antibody discovery. This demonstrates the capability of the 1-read-1-cell principle in conducting two-target analyses of mammalian cells and potentially even more than two targets. Therefore, the use of barcoding-free high-throughput sequencing with the 1-read-1-cell principle allows for the efficient processing of large numbers of cells and concurrent analysis of multiple targets.


