Single-cell assay for transposase-accessible chromatin by sequencing (scATAC-seq) is a technique employed to evaluate the accessibility of chromatin across the entire genome. It reveals the genomic regions responsible for gene regulation, offering profound insights into the mechanisms controlling cellular development, differentiation, and responses to stimuli, perturbations, or disease. Several open-source and commercial scATAC-seq methods have been developed, enabling the design of large single-cell atlas studies. However, to date, there has been a lack of comprehensive benchmarking for these methods, preventing meaningful comparisons between datasets generated by different techniques and hindering informed technology selection. In a recent publication in Nature Biotechnology, De Rop et al. report the outcomes of a multicenter benchmarking study of eight scATAC-seq protocols, marking the first comprehensive evaluation of this technology. Additionally, they introduce PUMATAC, a versatile preprocessing pipeline that can accommodate the diverse sequencing data formats produced by these methodologies.
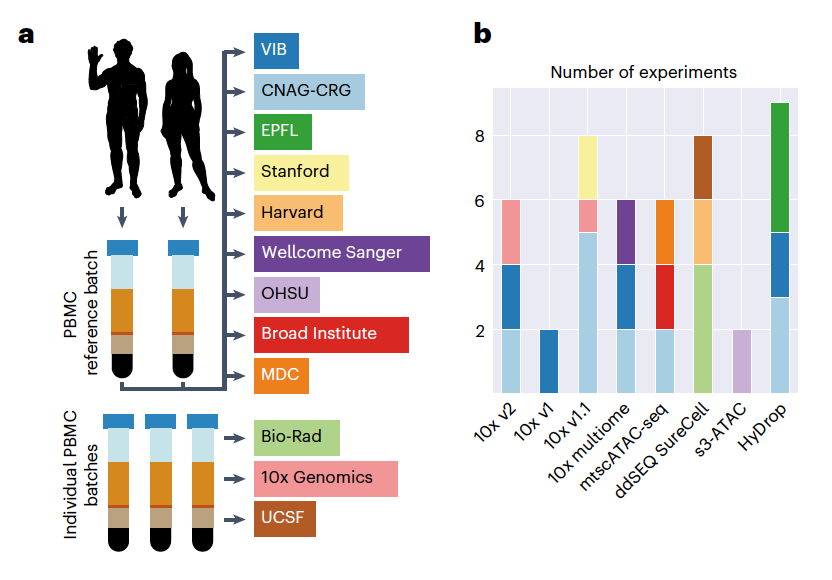
Twelve different institutions participated in the systematic benchmarking of 8 scATAC-seq protocols, which included the open-source HyDrop and s3-ATAC protocols, all variants of 10x Genomics scATAC-seq (v1, v1.1, v2, mtscATAC), and Bio-Rad ddSEQ. To limit variability to the tested protocols, human peripheral blood mononuclear cells (PBMCs) were used as reference samples by all institutions. Technical replicates were performed for each experiment, and 47 datasets were produced. For standardized comparison, all sequencing data were processed using PUMATAC, a newly developed scATAC-seq preprocessing pipeline that includes cell barcode error correction, adapter trimming, reference genome alignment, and mapping quality filtering. After initial preprocessing, cisTopic was used to identify and remove background noise barcodes and low-quality cells from the sample. To ensure consistency, all datasets were downsized to approximately 40,000 (40k) reads per cell. This dataset, or subsets derived thereof, was used for all downstream analyses.
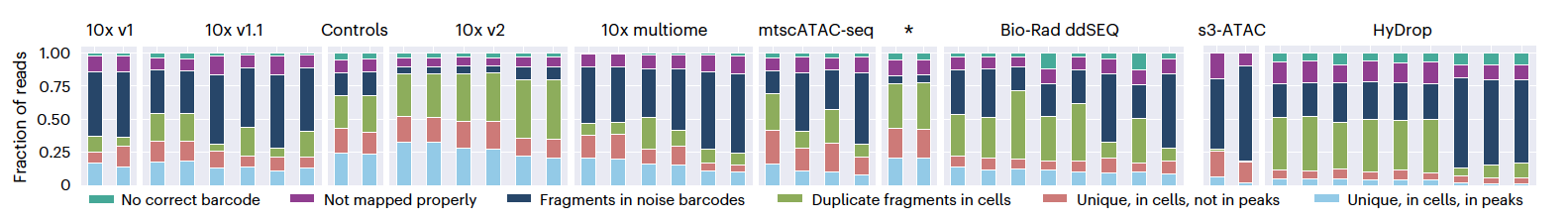
As the PUMATAC pipeline discards sequencing reads at several phases throughout preprocessing, the fraction of total reads lost at each filtering stage was calculated to determine sequencing efficiency and to shed light on the causes of read loss. This calculation demonstrated that sequencing efficiency in scATAC-seq experiments is generally low, ranging between 4-28%, with a significant fraction of original sequencing reads being discarded during preprocessing. The open-source protocols suffered the largest losses, although this was due to distinct mechanisms: s3-ATAC samples contained many fragments outside of peak regions, whereas HyDrop samples had highly duplicated fragments. Given that these protocols have not undergone rigorous optimization and are open-source, however, there is potential for further protocol refinement to enhance cell quality and library complexity and to reduce issues like ambient chromatin contamination and PCR duplication through community-driven improvements.
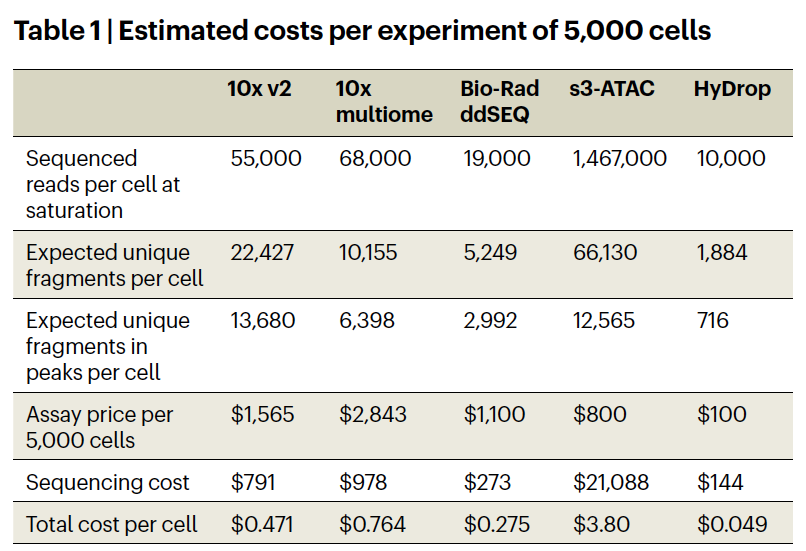
After the initial filtering steps, there were significant variations in quality metrics among the remaining cells, highlighting notable distinctions between open-source and commercial methods. Generally, the open-source protocols demonstrated lower sensitivity and poorer performance in most quality control assessments compared to their commercial counterparts. While all methods generally agreed on cell-type identity and transcription factor activities, the differences in sensitivity were evident when analyzing rare cell types, which showed lower assignment scores across techniques. The open-source protocols recovered fewer or no cells in these cases, indicating the need for increased sensitivity to achieve comprehensive cell-type annotation. However, when planning large-scale experiments, it is also important to consider cost-effectiveness in the decision-making process, as open-source protocols offer a substantial cost advantage (5x-10x cheaper per cell) over commercial assays. Therefore, it is crucial to weigh the higher reagent costs associated with commercial methods against the trade-off in accuracy seen in open-source variants. Additional factors such as method accessibility and compatibility with diverse datasets across studies should be key considerations influencing technology selection.


