Industrial enzyme preparations largely rely on microbes because of their ability to produce enzymes in bulk at a low cost, eco-friendly nature, ease of handling, and adaptability for product modification. Strategies incorporating genetic and process engineering have been employed to enhance microbial strain development for increased productivity, stress tolerance to production conditions, and improved yields due to increasing demands in the industry. However, these approaches are limited due to the complex nature of metabolic processes and our incomplete understanding of their mechanisms. Additionally, the use of recombinant DNA technology in the food industry is highly regulated, and there is generally negative feedback from consumers towards genetically modified food ingredients. Directed evolution using random mutagenesis and high-throughput screening (HTS) can overcome these limitations. In this study, an integrated microfluidic droplet sorting platform based on fluorescence-activated droplet sorting (FADS) was used to screen a mutant library of α-amylase-producing Bacillus licheniformis mutants generated by atmospheric and room temperature plasma (ARTP). Mutants with up to 50% higher α-amylase production were successfully identified, suggesting the utility of this approach for future strain development.
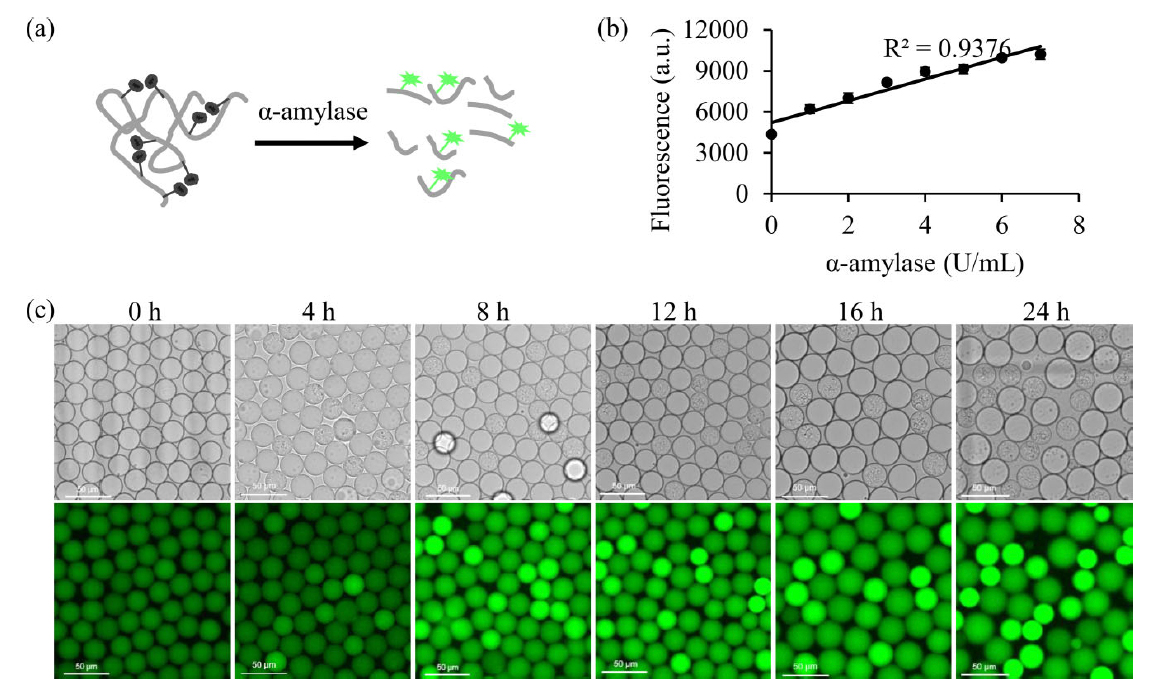
In this study, the authors used BODIPY fluorescein-labeled starch substrate to measure α-amylase activity of mutants encapsulated in droplets. As amylases hydrolyze the starch backbone, fluorophores are released, generating a fluorescent signal for imaging and droplet sorting. The production of α-amylase was found to be proportional to the increase in fluorescence intensity in wild-type B. licheniformis (R2=0.9376). Additionally, these reactions could be monitored over a 24-hour period with microscopy images demonstrating that fluorescence intensities gradually increased over time. Therefore, the BODIPY FL-labeled starch could directly measure concentration of α-amylase. From this experiment, the authors also chose to continue with 8-hour incubation time for screening experiments to allow adequate secretion of enzyme for screening and avoid overabundant bacterial growth.
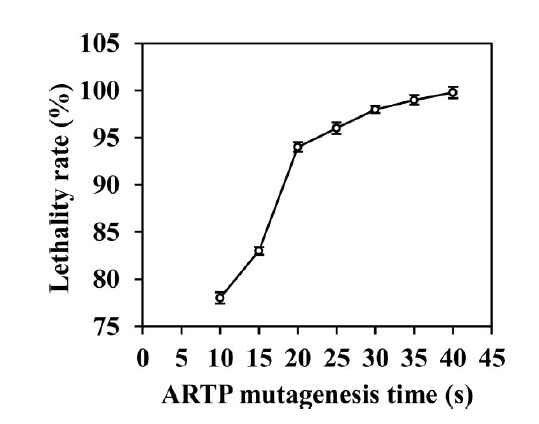
The authors first had to generate a library of B. licheniformis mutants prior to screening for high α-amylase production strains. This was done using atmospheric and room temperature (ARTP) mutagenesis. A single colony of B. licheniformis was grown to logarithmic phase and ten microliters of the culture was treated with ARTP for 0-45 seconds. Following mutagenesis, cells were washed and plated on LB to check for lethality. An exposure time higher than 20 seconds was chosen for subsequent experiments because a lethality rate between 90-95% resulted in the highest positive mutation probability. To increase genetic diversity of the mutant library, the authors repeated this experiment ten times and then combined all recovered mutants. Mutant strains were then encapsulated and sorted using the FADS platform. A total of ∼648 droplets (0.03%) were sorted from 2.1 × 106 droplets within 2 hours. The sorted droplets were then broken, and mutants were cultured on LB plates for subsequent fermentation experiments.
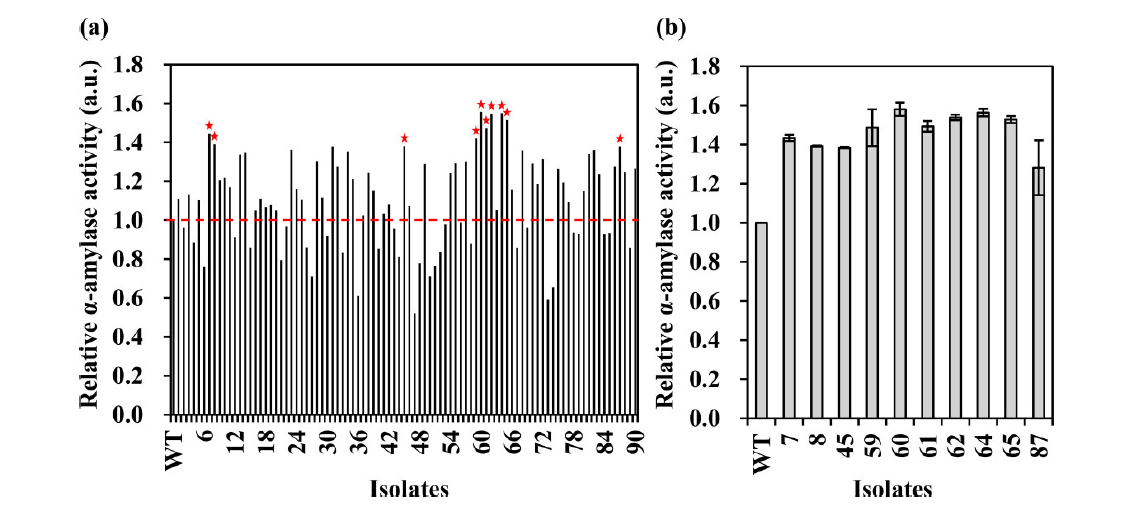
From the mutant library, 90 colonies were randomly picked and analyzed for their α-amylase production in shake-flask fermentation. Over half of the 90 mutant colonies screened (64.4%) exhibited higher enzyme production than the wild-type strain, and the top-performing clone had a more than 50% increase in α-amylase activity. To ensure that the genetic mutations induced by ARTP treatment were stable, the authors screened the 10 highest yield α-amylase producing strains in a secondary experiment. Results demonstrated that 90% of the mutants maintained the same level of enzyme production, suggesting that the mutants were stable. Overall, these results illustrated that droplet microfluidic HTS platforms can accelerate the process of directed evolution. Compared to conventional microtiter plate-based screening formats, the FADS platform could shorten the entire screening process from weeks to hours while reducing the costs of reagents and consumables. The platform could be adapted to screen microbes for the production of other enzymes or metabolites.


