Recent developments in multi-omics technologies have spurred renewed research interest in resolving the complex contributions of the human gut microbiota to the maintenance of health and development of disease. Among the myriad chronic diseases that have been linked with gut dysbiosis, such as rheumatoid arthritis, inflammatory bowel disease, and asthma, it has also been established that differences in the composition and function of gut microbiota cause variations in energy metabolism, fatty acid tissue composition, and low-grade inflammation; these are factors that play a role in obesity, which currently affects 650 million adults and 124 million children worldwide. The mechanism linking gut microbiota to obesity development is still under investigation; nonetheless, numerous studies are examining the possible use of engineered bacteria for obesity therapeutics. In this study, Yin, Chen, and others from the Frontiers Science Center for Synthetic Biology in Tianjin, China, build on previous work from their group in which they engineered butyrate-producing bacteria that had anti-obesogenic effects on mice. Here, they develop a droplet-assisted method to isolate and enrich functional gut bacteria from faecal bacterial suspensions that can use butyrate or other metabolites from this engineered strain. These metabolically functional species can be used for future investigations of bacterial therapeutics for the treatment of obesity.
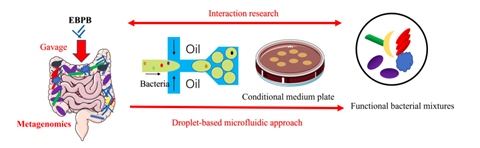
In their previous work, the authors constructed an engineered butyrate-producing bacteria (EBPB) using Bacillus subtilis. Butyrate has been shown to promote the growth of beneficial intestinal microbiota, provide energy to colonic epithelial cells, regulate energy absorption, improve glucose homeostasis, and increase insulin sensitivity. When administered orally, butyrate compounds have low bioavailability and are not effective in treating obesity. However, ingestion of EBPB significantly inhibited weight gain and insulin resistance in mice fed a high-fat diet; it also resulted in an increase in the abundance of beneficial bacterial species, such as Bifidobacterium, Lactobacillus, and Akkermansia, as revealed by metagenomic sequencing. Despite knowing the genetic identity of these beneficial strains that may interact with EBPB, the authors have been unable to successfully culture these bacteria for further study, in part due to competition with fast-growing bacterial species on culture plates. Thus, they sought to develop a novel culture approach using droplet-based microfluidics, as single-cell encapsulation provides a protective environment for cells to grow without competition from others. In addition, the inherent scalability of a droplet-based method allows thousands of such droplets to be cultured in parallel, greatly enhancing throughput. And, since each droplet serves as a microenvironment for the single cell inside, it can be fine-tuned to reflect the needs of each species; for example, the surrounding oil can be pre-treated to generate an anaerobic environment. As a result, a droplet-based culturing approach that could interface with traditional culture methods and metagenomic studies is highly desirable.
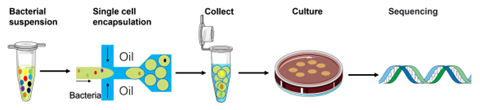
To develop their single-cell droplet culture approach, the authors first utilized a bacterial suspension containing Escherichia coli Nissle 1917, known to have probiotic properties and a relatively rapid growth rate, and Bifidobacterium pseudocatenulatum, which is also known to have probiotic properties but is strictly anaerobic and difficult to culture on agar plates. Cells were loaded onto the microfluidic device at a density that ensured single-cell encapsulation, with the percentage of single-cell droplets near the theoretical value of 1%. Individual droplets were then spread onto an agar plate, where each cell could culture independently for approximately 8 hours before the droplets broke, whereby the cells could continue to culture on the agar plate; each colony on the plate would thus be derived from a single cell. Compared to serial dilution, this method of achieving colonies from single cells offers the advantages of simplicity and speed. Importantly, the droplet-based method was singularly capable of producing colonies of both species, whereas traditional culture methods only resulted in colonies of E. coli. This suggests that the droplet-based culture method can enhance cell growth, especially for slow-growing species such as anaerobic gut bacteria.
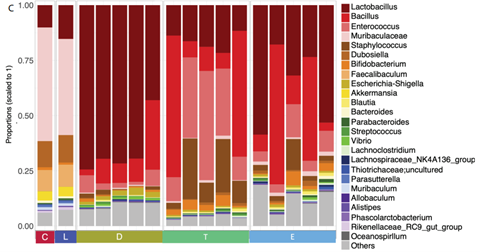
The droplet-based culture method was then repeated using a pre-filtered faecal bacterial suspension. Compared to traditional cultivation methods, the plates formed via the single-cell droplet approach contained more colonies. Analysis of 16s rRNA sequencing data revealed that there was no significant difference between the alpha diversity (richness) of the cells from the original faecal sample and the cells from the droplet culture, suggesting that the communities obtained from the droplet culture method are the same as the communities present in the original sample. Beta diversity, which considers community composition, differed between traditional culture plates and droplet-based culture plates, with most of the top-25 genera increasing in abundance in the droplet-based method. In addition, within a random selection of 100 colonies, a total of 7 species were found in traditional culture plates, whereas a total of 14 species were found in droplet culture plates, which suggests that the single-cell droplet culture is more effective for culturing gut microbiota and preserving community diversity. In a final experiment, the authors spread droplets generated from the original faecal suspension onto plates containing the supernatant of EBPB, which contains butyrate and other metabolites. Beta-diversity analysis suggested that the resulting community was unique, with 11 species under four genera (Lactobacillus, Bifidobacterium, Bacillus, and Enterococcus) present, which represent species able to use butyrate and/or other EBPB metabolites. Thus, the droplet-based culture method successfully obtained functional gut bacteria isolates to be used in future studies of anti-obesity bacterial therapeutics.


