The efficient secretion of heterologous proteins by engineered bacterial host cells has both therapeutic and commercial applications, such as in the production of antibodies, growth factors, nanobodies, and high-quality protein food ingredients. Metabolic engineering of bacterial hosts generally involves rational design, in which gene regulatory elements or synthetic gene circuits are engineered to produce desired gene products in cell factories. However, challenges remain in terms of engineering bacterial host strains for optimal protein secretion, which may be alleviated through complementary use of a directed evolution scheme. To that end, the authors of this study use random mutagenesis followed by high-throughput droplet-assisted screening to identify Corynebacterium glutamicum strains with favorable protein secretion phenotypes. They also investigate the resulting mutant library for favorable mutations that could benefit rational engineering strategies in the future.
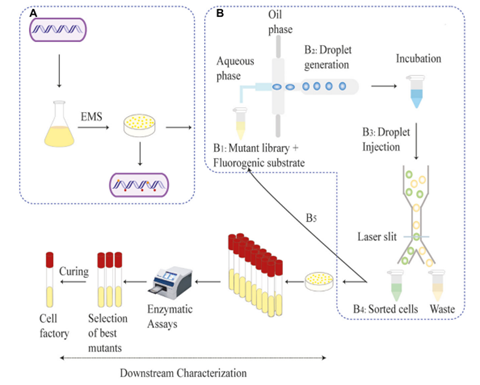
The C. glutamicum strain JS34 was selected as the bacterial host for strain construction. This gram-positive soil bacterium is already widely used in industrial applications, has low nutrient requirements, lacks extracellular proteases, and is denoted GRAS (generally regarded as safe) by the United States Food and Drug Administration. It has also been used to produce pharmaceutical products such as vitamins and amino acids. A plasmid containing the β-glucosidase gene from S. degradans, the SD sequence of the tpiA gene (ribosomal binding site upstream of the start codon), and the signal peptide Cg0949 was constructed and transformed into C. glutamicum JS34, resulting in strain JC0190 capable of secreting β-glucosidase. This strain was mutagenized with ethyl methanesulfonate for 3 hours, which resulted in a 99.68% killing rate. The surviving mutant cells were encapsulated as single cells in droplets with Fluorescein Di-β-DGlucopyranoside (FDGlu), which fluoresces green after being hydrolyzed by β-glucosidase. The droplets were incubated for 3-4 hours, allowing time for β-glucosidase secretion, and then sorted based on the intensity of the fluorescent signals. After two rounds of screening, the 3,900 droplets with the highest fluorescent signals, representing 1.95% of total positive droplets, were selected. The bacterial cells were released from the droplets and recovered on agar plates. A secondary screening, whereby β-glucosidase-producing colonies would produce a brown color on the agar plates, was used to select the sixteen colonies with the greatest capacity for protein secretion. These sixteen strains were subjected to further analysis.
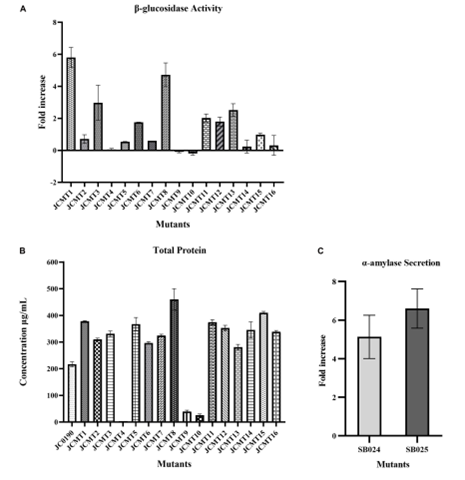
First, the β-glucosidase activity in the supernatant was measured via hydrolysis of FDGlu. Mutant strain JCMT1 had sixfold higher β-glucosidase activity compared to control strain JC0190; mutant strain JCMT8 had fivefold higher activity compared to control. The supernatants of all sixteen mutant strains were then concentrated and protein secretion was analyzed on an SDS-PAGE gel and quantified using the Bradford assay. Mutant strain JCMT8 had the highest total protein content in the supernatant; mutant strain JCMT1 had the third highest. JCMT8 also had the highest biomass production among the mutant strains. Both JCMT8 and JCMT1 were thus selected as high-performing mutants and tested for their ability to produce proteins other than β-glucosidase. To do this, plasmid curing was used to eliminate the β-glucosidase production plasmid from the bacterial hosts. The JCMT8 and JCMT1 mutants were then transformed with a new plasmid containing the α-amylase gene from G. stearothermophilus, the SD sequence of the tpiA gene, and the signal peptide Cg0949. The resulting strains maintained similar growth rates and produced 5.6-7x more α-amylase than the control. This suggests that both mutant strains can efficiently produce heterologous proteins, and that protein secretion is improved compared to the parent control strains. As a result, these strains could serve as efficient cell factories for industrial protein production.
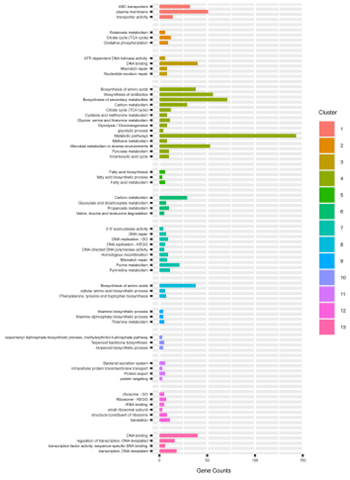
To better understand the genotypic changes that resulted in the improved protein production seen in the mutant strains, the genomes of all 16 mutant strains, along with that of the parent strain JC0190, were sequenced. Between 50-150 unique single-nucleotide variants (SNVs) were found in each of the 13 mutant strains with unique SNV profiles compared to the parent strain, which represented an occurrence of 0.002-0.005% in the genome and 0-0.012% in the plasmid. Across these 13 mutants, the authors identified 313 silent mutations, 566 missense mutations and 123 mutations in the non-coding region, for a total of 1002 SNVs. Functional enrichment and clustering analysis resulted in 13 functional clusters, relating to membrane transport activity, primary and secondary metabolisms including the citric acid cycle and glycolysis, protein secretion, and translation and ribosome processes, among other terms. The analysis of these mutations provides valuable starting points for future rational design of bacterial host cells as protein-producing cell factories.


