Microfluidic platforms have revolutionized the way biochemical experimentation is conducted. As versatile and highly efficient catalysts, enzymes play a crucial role in initiating, accelerating, and controlling a wide range of reactions in living systems. These enzymes have evolved over time, optimizing their abilities to ensure precise substrate specificity and remarkable selectivity. However, despite their structural and chemical diversity, only a limited number of natural enzymes have been effectively employed in industrial settings. One practical solution lies in microfluidic droplets, which resemble cells in terms of size and isolated environments. These droplets provide a practical platform for conducting enzyme-catalyzed reactions in vitro, while preserving their high catalytic activity. The applications of enzyme catalysis in microfluidic droplets are wide-ranging. From studying fundamental enzymatic mechanisms to developing novel biocatalytic processes, researchers are exploring the potential of these platforms in diverse fields such as drug discovery, biofuel production, and synthetic biology.
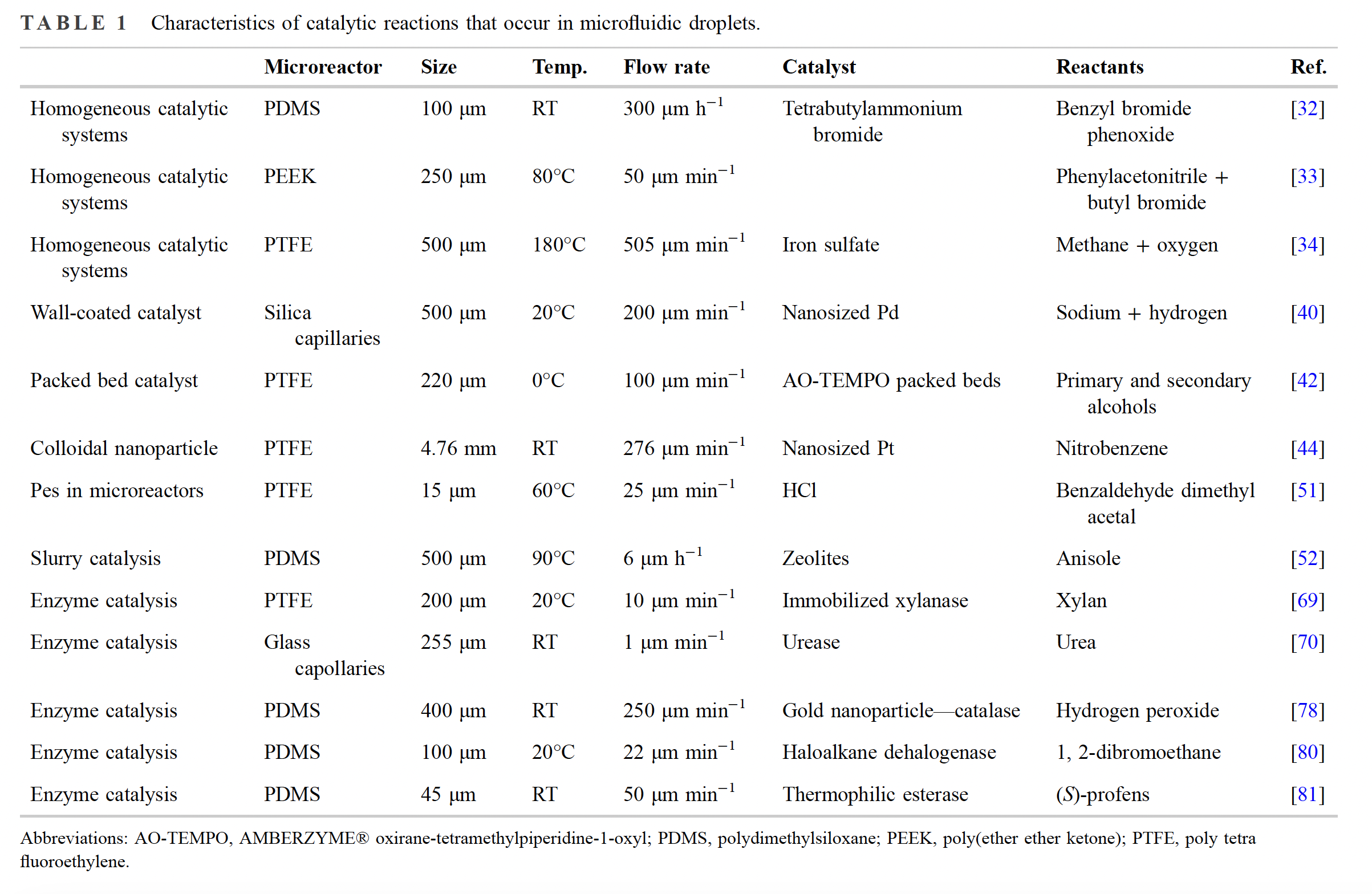
Enzyme catalysis differs from traditional metal and small molecule-based catalysis in its remarkable specificity. The selectivity of enzyme catalysis depends on the binding to specific sites on the substrate. Enzyme-catalyzed reactions exhibit cascaded amplification under specific stirring time and solution environments, similar to the polymerase chain reaction. Enzymatic catalysis in organisms is often confined to the internal environment of cells, bacteria, or viruses, which significantly differs from the open solution environment. There are several advantages of utilizing microfluidic droplets for enzyme catalysis. Firstly, the small droplet size (ranging from picoliters to nanoliters) ensures a large surface-to-volume ratio, facilitating efficient mass transfer and rapid reaction kinetics. Moreover, the compartmentalization provided by the droplets allows for the isolation of individual reactions, minimizing cross-contamination and enabling parallel screening of multiple enzyme variants or conditions. The ability to precisely manipulate droplets also allows for easy mixing, concentration gradients, and the introduction of multiple reactants, enabling the study of complex reaction networks. Finally, microfluidic droplets offer the opportunity to investigate enzyme kinetics at unprecedented spatiotemporal resolutions. Users can monitor reaction progress in real-time by incorporating fluorescent probes or reporters into the droplets, gaining valuable insights into reaction dynamics and heterogeneity. Therefore, droplet microfluidic systems enable high-throughput screening of enzyme variants, leading to accelerated enzyme engineering and discovery in an unbiased manner.

In one example, Ma et al. used a dual-channel microfluidic droplet screening system for efficient molecular evolution to generate enantioselective profens. Profens, utilized for treating pain, inflammation, fever, and stiffness, are chiral molecules. The therapeutic properties of profens drugs are attributed to the (S)-enantiomers, whereas the (R)-profens have the potential to induce severe side effects by interfering with normal lipid metabolism and membrane function. Therefore, it is highly desirable to generate (S)-profens. Traditional methods for enzyme evolution and screening can be time-consuming and labor-intensive. However, the dual-channel microfluidic droplet screening platform offers advantages such as high-throughput capabilities, precise control over reaction conditions, and the ability to screen a large number of enzyme variants in parallel. The authors were able to screen up to 107 Archaeoglobus fulgidus (AFEST) mutants that preferentially produce (S)-profens. To screen for droplets containing (S)-ibuprofen, the authors synthesized a set of fluorogenic substrates that selectively esterify (S)-ibuprofen or (R)-ibuprofen. Following five rounds of directed evolution, Ma et al. identified variants that exhibited 700-fold increased enantioselectivity for the desired (S)-ibuprofen. These results demonstrate the advantages of droplet microfluidic in the rapid generation and identification of enzymes with desired properties.

In another example, Hess et al. introduced a droplet microfluidic system for the rapid measurement of multiple enzyme reactions. The droplets were manipulated by controlling their velocity, directing them through a serpentine channel path to induce rapid acceleration and facilitate swift mixing. The oxidation of Ampliflu Red (AR) mediated by ferric microperoxidase (MP-11) and the hydrolysis of resorufin β-D-galactopyranoside (RGP) mediated by β-galactosidase were employed to investigate transient kinetics and thermodynamics. The results of kinetics analysis revealed that resorcinol oxidation occurred rapidly, while the formation of resazurin was displayed slower kinetics. The glycosidase hydrolysis exhibited a linear reaction, indicating a lack of equilibrium under the experimental conditions. Furthermore, by encapsulating these reactions in droplets, the assay volumes reduced by six orders of magnitude while increasing screening to 9,000 reactions/min. These findings demonstrate that droplet microfluidics enabled efficient and systematic acquisition of comprehensive kinetic data for in-depth exploration of enzyme catalysis. The study's findings provide novel insights into substrate specificity at the molecular level and shed light on the role of hydration-related entropy through the integration of high-throughput data collection and global molecular dynamics simulations.
- Mei F, Lin H, Hu L, Dou WT, Yang HB, Xu L. Homogenous, heterogenous, and enzyme catalysis in microfluidics droplets. Smart Molecules. 2023 Apr 17; e20220001.
- Ma F, Chung MT, Yao Y, Nidetz R, Lee LM, Liu AP, Feng Y, Kurabayashi K, Yang GY. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nat Commun. 2018 Mar 12;9(1):1030. doi: 10.1038/s41467-018-03492-6. PMID: 29531246; PMCID: PMC5847605.
- Hess D, Docklova V, Kokkonen P, Bednar D, Damborsky J, DeMello A, Prokop Z, Stavrakis S. Exploring mechanism of enzyme catalysis by on-chip transient kinetics coupled with global data analysis and molecular modeling. Chem. 2021 April 8;7(4):1066-1079.


