Microorganisms like the baker's yeast Saccharomyces cerevisiae serve as valuable platforms for heterologous protein production, prompting extensive commercial and scientific endeavors to enhance the efficiency and yield of protein production and secretion in these organisms. However, the intricate and multifaceted nature of these processes, which involve a multitude of components and pathways, necessitates the use of high-throughput screening techniques to pinpoint the specific bottlenecks and areas that warrant enhancement. In this context, the integration of high-throughput droplet microfluidic screening with next-generation sequencing is particularly advantageous, as it offers a precise and comprehensive approach to systematically explore the effects of gene perturbations on protein secretion and production. In this study, Johansson et al generate massive CRISPR activation and repression libraries to alter the expression of more than 7000 genes and ncRNAs in S. cervisiae, marking a significant step towards comprehensively probing the cellular machinery. Using droplet microfluidics, they conduct high-throughput functional screening of this variant library to examine which of these genetic changes had a discernible impact on α-amylase production.
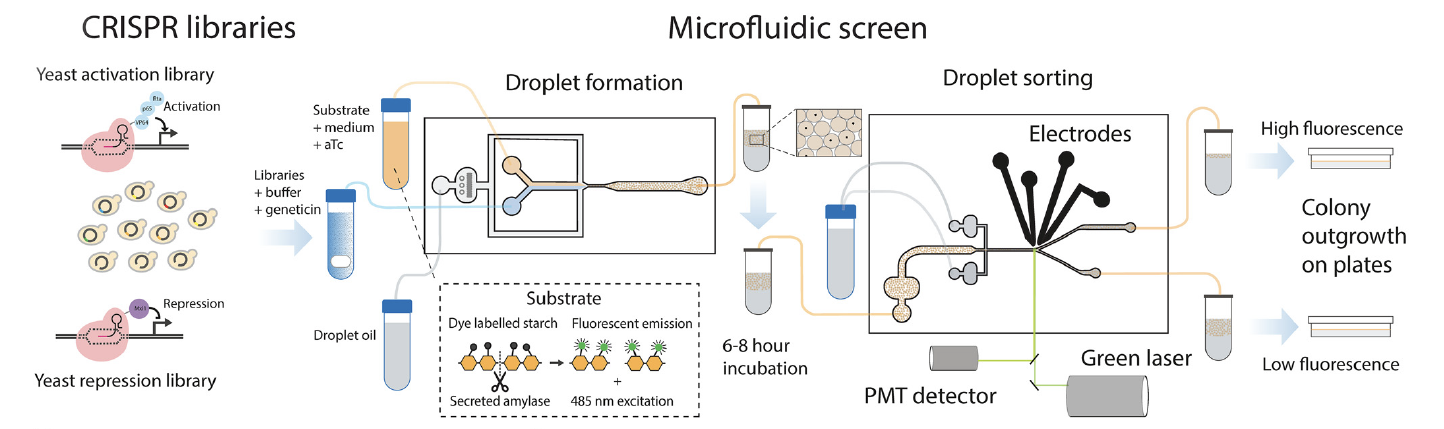
The yeast strain chosen for this study was Ethanol Red, commonly used for bioethanol production, which was genetically modified to express α-amylase. The modified strain was transformed with plasmid libraries for gene activation or repression, which had been generated using CRISPRa and CRISPRi, respectively. The resulting cells with plasmid libraries were encapsulated in droplets along with growth medium, the fluorescent substrate BODIPY–starch, and an inducer for guide RNA expression, using a flow-focusing droplet generator chip. The droplets were incubated to allow for accumulation of secreted α-amylase. As α-amylase concentration increases within the droplets, the enzyme hydrolyzes the BODIPY-starch substrate, which results in an increase in fluorescence. Thus, fluorescence-activated droplet sorting (FADS) can be used to conduct a high-throughput functional screen of α-amylase activity. The sorting gates were chosen to select the top 2-5% of droplets based on fluorescence. These droplets were collected and broken, and the resulting cells were plated on YPSD geneticin plates for overnight growth. The plasmid guide regions then underwent PCR amplification, and next-generation sequencing of the amplicons was used to identify which genes were most strongly associated with the observed increase in protein secretion.
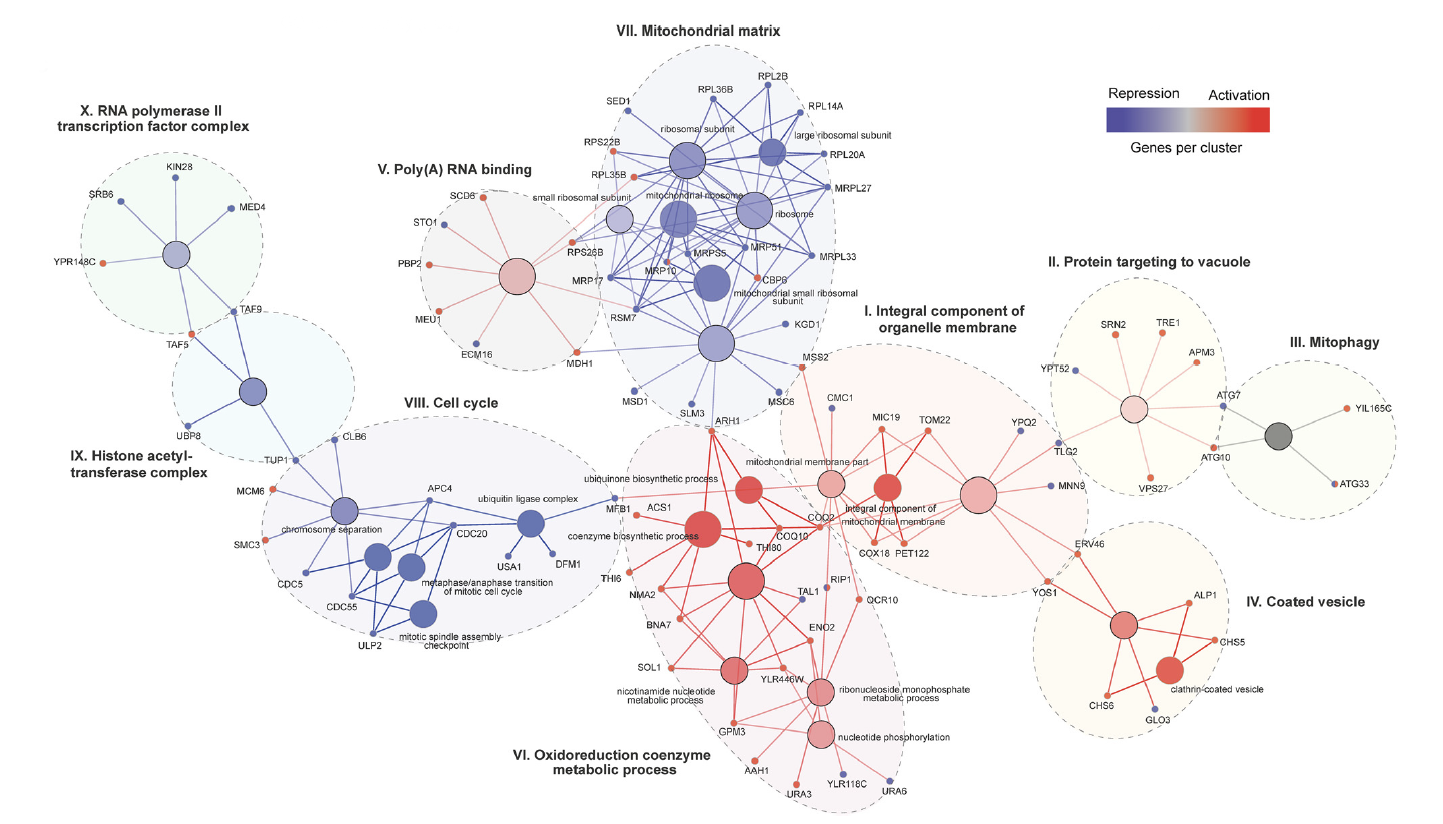
Two million droplets were screened using FADS, achieving 20-fold library coverage. In total, 11,218 guides from activation screens and 20,561 guides from repression screens were identified, providing 83% gene coverage for the activation screens and 97% gene coverage for the repression screens. Within the sorted droplets, bioinformatics analysis pinpointed 345 genes that, when modulated in expression, resulted in an increase in α-amylase secretion. GO enrichment analysis was employed to identify the specific cellular processes and pathways associated with these genes, then data from both the activation and repression screens were integrated into a single map of genes found to increase α-amylase production. This map consisted of 10 clusters, six of which were composed primarily of genes from the activation screens. These clusters highlighted the role of genes related to diverse cellular processes such as mitochondrial function, vesicle trafficking, RNA binding, transport, ribosomal subunits, cell cycle regulation, and transcriptional processes in modulating α-amylase secretion. In general, elevating gene expression related to secretory pathway components, while repressing cell cycle and ribosomal activity genes led to higher α-amylase secretion. In vitro validation of enriched genes from the droplet-based functional screens confirmed that these changes result in 20-30% increases in α-amylase secretion that were sustained over 24 and 48 h.
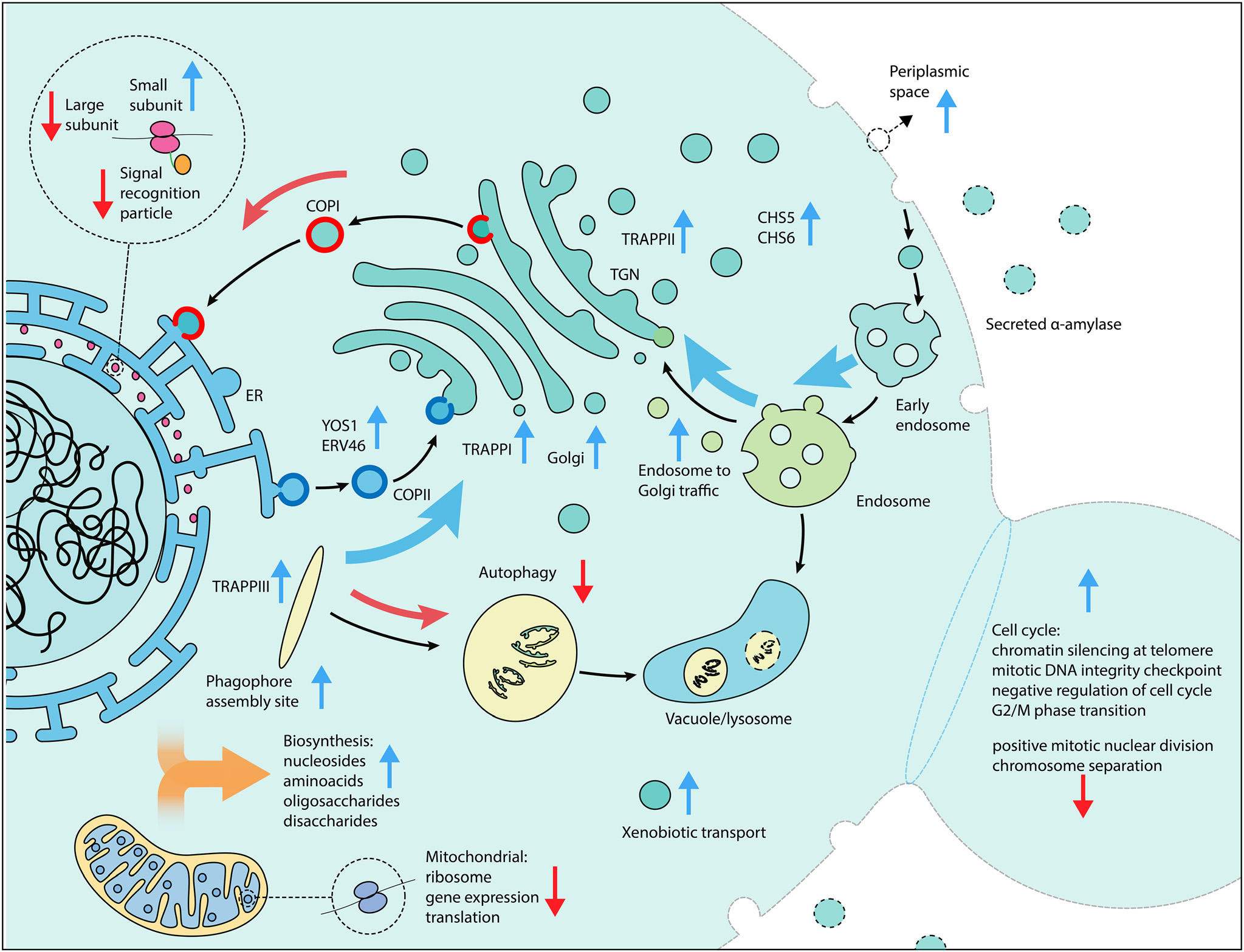
Non-coding RNAs, including 71 stable unannotated transcripts (SUTs) and 8 cryptic unstable transcripts (CUTs), were found to be enriched in both the activation and repression screens, suggesting their potential involvement in modulating α-amylase secretion. Enrichment analysis revealed their proximity to genes associated with cellular processes related to vacuoles, endosomes, and the Golgi apparatus. These non-coding RNAs may serve as bidirectional promoters, potentially activating target genes, while others could overlap or interfere with the transcription of their antisense counterparts, resulting in reduced expression. Further experimental investigations are required to fully uncover the roles of these non-coding RNAs in α-amylase secretion regulation. The study's comprehensive design, which identified and validated novel gene targets and potential regulatory elements for α-amylase secretion, in conjunction with the precise and efficient protocol, holds promise as a blueprint for future large-scale investigations. It enables the swift evaluation of gene expression changes and could be used to promote iterative strain enhancements, providing valuable insights for biotechnological applications.
- Johansson SA, Dulermo T, Jann C, Smith JD, Pryszlak A, Pignede G, Schraivogel D, Colavizza D, Desfougères T, Rave C, Farwick A, Merten CA, Roy KR, Wei W, Steinmetz LM. Large scale microfluidic CRISPR screening for increased amylase secretion in yeast. Lab Chip. 2023 Aug 8;23(16):3704-3715. doi: 10.1039/d3lc00111c. PMID: 37483015; PMCID: PMC7614956


