The biological diversity of Earth is overwhelmingly microbial. Since the development of cultivation-independent molecular analysis techniques, we have witnessed a burst in the detection of previously elusive microbial taxa. The analysis of all DNA from a bulk sample, known as metagenomics, has become a widely adopted approach to obtaining hundreds or thousands of genomes from environmental samples. Alternatively, single-cell genomics has emerged as a technique to recover genomic information from individual cells and shed light on a strain-level diversity of complex cell populations. This simplifies challenges associated with metagenome assembly and provides a direct link between the genome and any additional DNA, such as phages or plasmids [1].
Depending on the research question at hand, single-cell sequencing can follow two primary approaches: whole-genome sequencing or targeted sequencing. The latter option is particularly attractive for hypothesis-driven research involving the study of thousands to millions of individual cells. Specific primers can be designed to target desired 16S rRNA genes, biosynthetic gene clusters, drug-resistant genes, and other regions of interest. A notable example of targeted sequencing applied for the characterization of functional diversity of complex microbial community on a single-cell level is an epicPCR (Emulsion, Paired Isolation and Concatenation PCR) technique. The epicPCR workflow involves linking functional genes with phylogenetic markers utilizing fusion PCR [2]. Moreover, the detection and quantification of specific microbial targets can be performed without the sequencing readout. Regardless of the goal and chosen analytical method, targeted detection requires a robust PCR assay.
Ultra-high-throughput analysis of tens of thousands of individual cells calls for cell partitioning into miniature compartments, and droplet microfluidics remains a preferred choice. While droplet-based approaches are powerful, they suffer from a fundamental limitation: cells lysed in droplets produce high concentrations of cellular contaminants that inhibit downstream assays. Furthermore, microbial cell lysis requires harsh lysis conditions that are not compatible with enzymatic reactions. As a result, droplet-based PCR methods require complicated, multi-step droplet manipulations to remove reaction inhibitors, making assays difficult and less reliable [3]. Moreover, even a well-established droplet digital PCR (ddPCR) method struggles with environmental samples due to the prevalence of droplets with intermediate fluorescence which cannot be reliably assigned to either positive or negative droplet population. This effect may be caused by the presence of inhibitors, droplet coalescence, or features of template DNA [4].
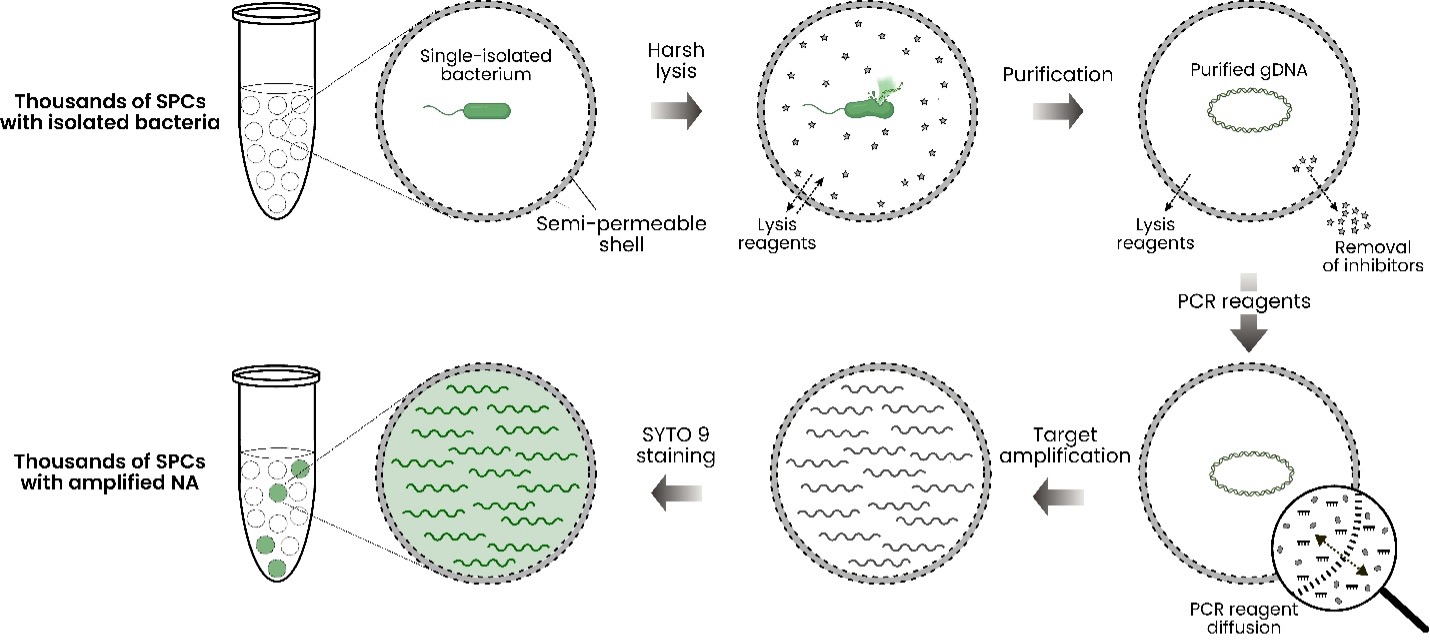
To demonstrate target amplification in semi-permeable capsules (SPCs), we encapsulated individual E. coli cells into SPCs on the Onyx platform, tested various PCR enzymes, characterized the retention of differently sized amplicons, and tested the effect of amplification occurring outside the compartments. The experimental scheme is shown in Fig. 1.
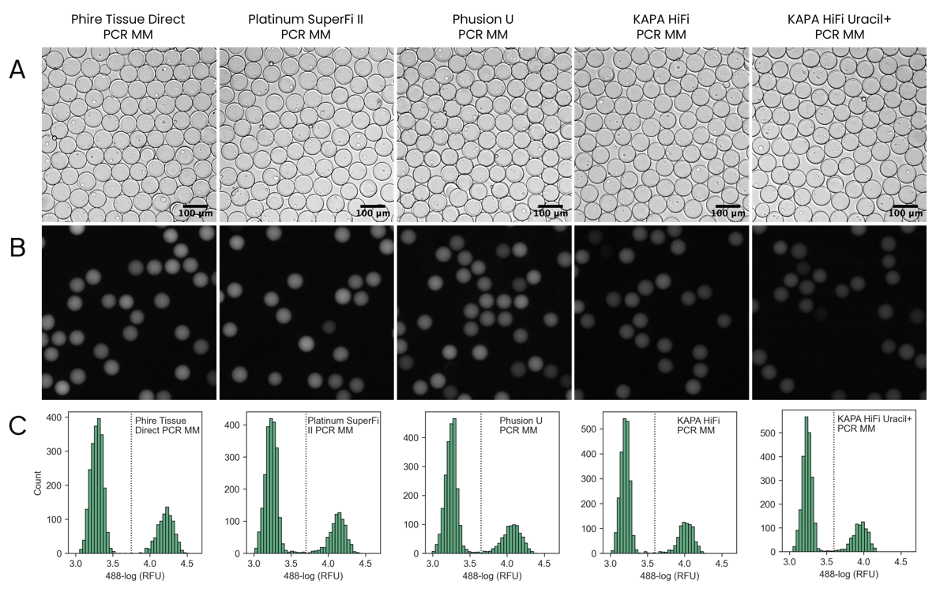
The results indicated that the best-performing PCR polymerase, as assessed by fluorescence microscopy, is the Phire Tissue Direct PCR Master Mix. For high-fidelity amplification, Platinum SuperFi II PCR Master Mix as well as the KAPA HiFi HotStart ReadyMix can be used, and uracil-containing templates may be successfully amplified by Phusion U Hot Start PCR Master Mix and KAPA HiFi HotStart Uracil+ ReadyMix (Fig. 2). Amplicon analysis by gel electrophoresis indicated specific amplification within SPCs in all cases.
Testing of various numbers of PCR cycles and amplicons of different lengths revealed the following: • PCR signal is detected starting with 25 cycles when the reaction is performed with Phire Tissue Direct PCR Master Mix and the amplicon size is ~500 bp. • PCR saturation is achieved in ≥ 35 cycles when the reaction is performed with Phire Tissue Direct PCR Master Mix and the amplicon size is ~500 bp. • The safe size limit for amplicon retention during PCR in SPCs is ≥ 500 bp. Note that given the nature of the test, this means 500 bp dsDNA and 500 nt ssDNA retention.
- 1. Bowers RM, Doud DFR, Woyke T. Analysis of single-cell genome sequences of bacteria and archaea. Emerg Top Life Sci. 2017;1(3):249-255.
- 2. Spencer SJ, Tamminen MV, Preheim SP, Guo MT, Briggs AW, Brito IL, A Weitz D, Pitkänen LK, Vigneault F, Juhani Virta MP, Alm EJ. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2016;10(2):427-36.
- 3. Hyman LB, Christopher CR, Romero PA. Single-cell nucleic acid profiling in droplets (SNAPD) enables high-throughput analysis of heterogeneous cell populations. Nucleic Acids Res. 2021;49(18):e103.
- 4. Kokkoris V, Vukicevich E, Richards A, Thomsen C, Hart MM. Challenges Using Droplet Digital PCR for Environmental Samples. Applied Microbiology. 2021; 1(1):74-88.


