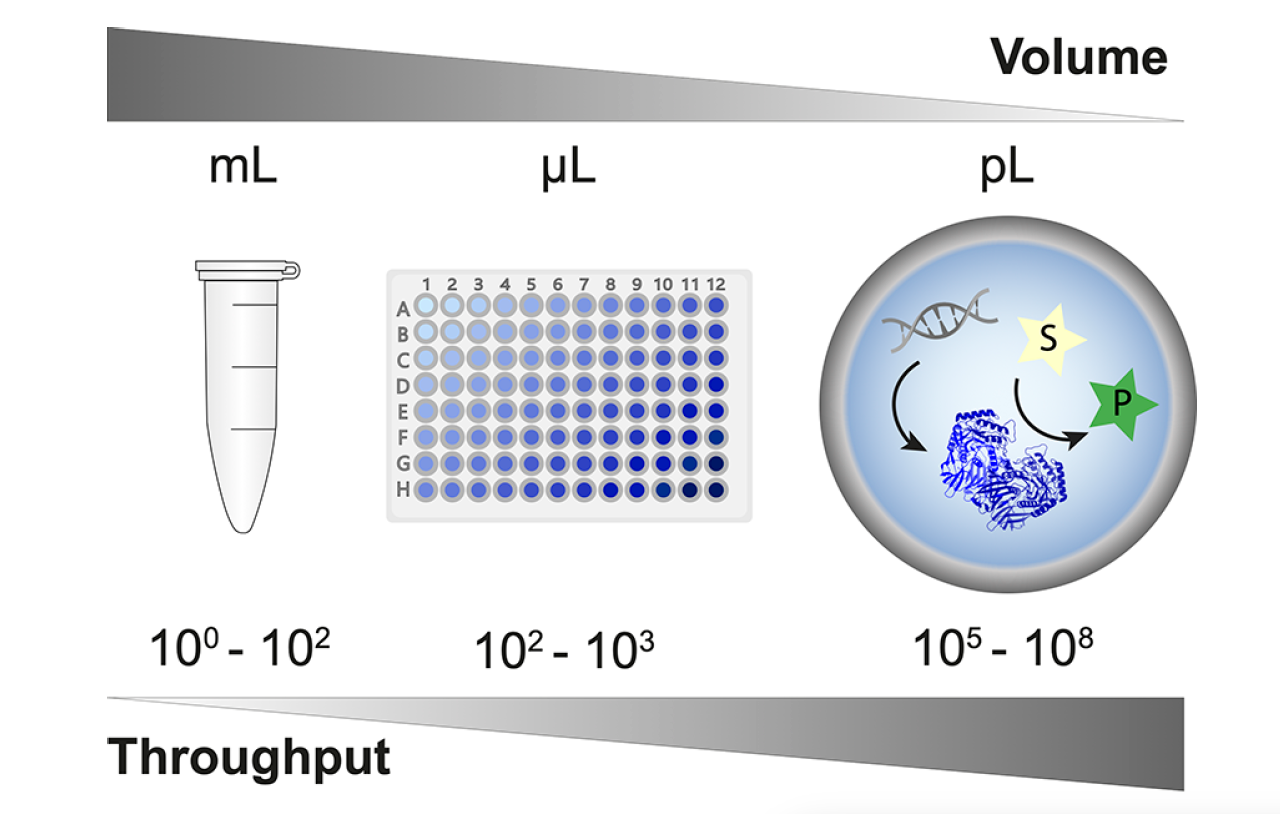
Droplet microfluidics technology, based on reaction miniaturization into picoliter- to nanoliter-sized compartments, is revolutionizing a wide range of diverse scientific disciplines by facilitating the ultra-high-throughput analysis of samples at the resolution of single cells and molecules. Within the field of synthetic biology, droplet microfluidics technology has been especially beneficial for protein engineering and enzyme screening campaigns, as the success of such campaigns often depends upon the number of protein variants tested. Given that the number of unique amino acid combinations can produce variant libraries reaching into the millions or higher, it is vitally important to employ the highest possible screening throughput, not only to avoid significant bottlenecks in the engineering design cycle but also to increase the likelihood of discovering rare and valuable candidates. Droplet microfluidics platforms, with droplet generation and sorting rates capable of analyzing millions of samples per day, are perfectly suited to navigating these ultra-high-throughput campaigns.
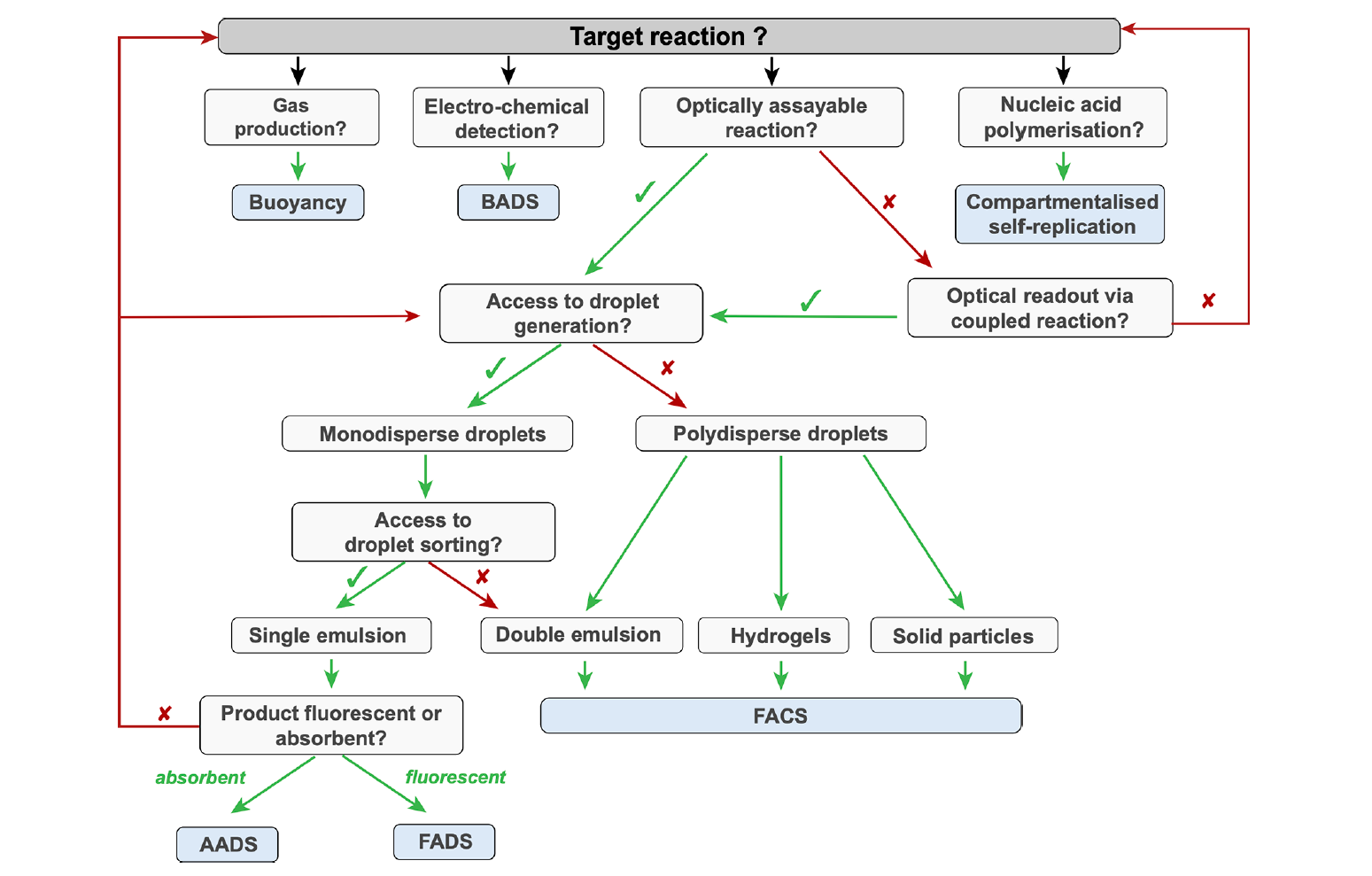
Successful high-throughput protein engineering campaigns using droplet microfluidics technology have already been demonstrated, and the development of droplet-based screening assays currently covers all seven enzyme commission number (EC) classes (oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases, and translocases). In a recent publication for Chemical Reviews, Gantz et al provide a comprehensive overview of droplet assays that have successfully been used in protein engineering campaigns. Some of these assays have been conducted using polydisperse emulsions, but the majority were conducted in monodisperse droplets created using a droplet generator. The utilization of a droplet generator to produce monodisperse emulsions ensures consistent droplet size and enables a more precise quantification of the reaction product using optical readout methods. Fluorescence-activated droplet sorting (FADS) is the most mature-on chip optical readout and is capable of sorting >107 droplets per day. It is compatible with virtually any assay where activity can be revealed by a fluorogenic substrate; this includes coupled assays, in which an optically inactive product can be converted into a downstream fluorescence signal.
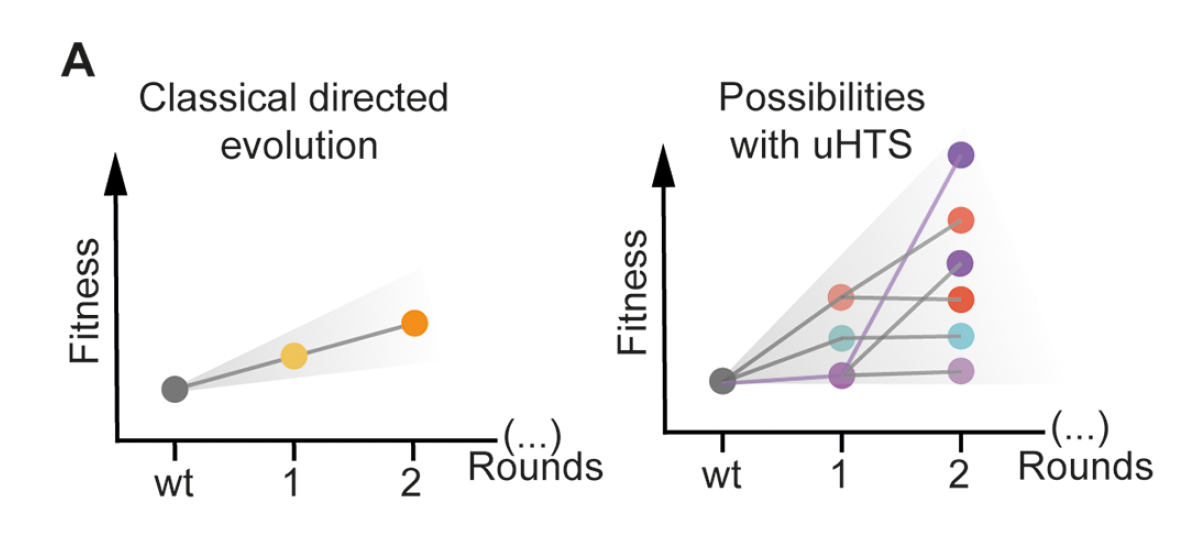
Notably, in many of the examples reviewed by Gantz et al, droplet assays not only accelerate the screening process but are fundamental to the success of the protein engineering campaign. This was the case for Obexer et al, who were able to rescue their stalled directed evolution campaign using droplet microfluidics. After five rounds of selection, their droplet-based directed evolution campaign improved the activity of an artificial aldolase 30x and yielded a completely remodeled active site. This exemplifies how increasing throughput can facilitate the exploration of more sequence space in the fitness landscape, thus increasing the likelihood of finding hits. Similarly, ultra-high-throughput droplet screening can be used to escape stalled evolutionary trajectories by offering opportunities to explore the synergistic interactions of mutations, introduce mutations that can bypass negative epistasis, or attempt more “inclusive” selection scenarios that tolerate multiple evolutionary trajectories in one experiment. As a result, ultra-high-throughput droplet screening changes the types of questions that can be asked and strategies that can be attempted during directed evolution campaigns.
Further, microdroplet-based ultrahigh-throughput screening of metagenomic libraries enables the discovery of functional information that defies prediction. For example, Colin et al used droplet microfluidics technology to screen a vast metagenomic library for phosphotriesterase activity, an uncommon hydrolytic reaction involving a non-natural substrate. Eight phosphotriesterases were discovered, including one that was a member of the α/β-hydrolase superfamily and exhibited esterase-like catalytic properties without the presence of an active site metal. Predicting promiscuous activities such as this is especially challenging, if not impossible, when using sequence-based methods. In addition, identifying hits for non-natural substrates is exceptionally rare. Statistically, a traditional robotic screen of tens of thousands of clones would have yielded a successful hit only once every ten attempts. The utilization of ultra-high-throughput droplet technology is therefore crucial in uncovering any hits in this context. As demonstrated by these examples, droplet microfluidics technology is driving a paradigm shift in protein engineering: uncovering functional information that defies prediction, providing a platform for exploring vast sequence spaces, and promoting the discovery of rare catalytic activities. Droplet microfluidics technology will redefine the boundaries of what is possible in protein engineering and pave the way for groundbreaking advancements in this field.
- Gantz M, Neun S, Medcalf EJ, van Vliet LD, Hollfelder F. Ultrahigh-Throughput Enzyme Engineering and Discovery in In Vitro Compartments. Chem Rev. 2023 May 10;123(9):5571-5611. doi: 10.1021/acs.chemrev.2c00910. Epub 2023 May 1. PMID: 37126602; PMCID: PMC10176489
- Obexer R, Godina A, Garrabou X, Mittl PR, Baker D, Griffiths AD, Hilvert D. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat Chem. 2017 Jan;9(1):50-56. doi: 10.1038/nchem.2596. Epub 2016 Aug 29. PMID: 27995916.
- Colin PY, Kintses B, Gielen F, Miton CM, Fischer G, Mohamed MF, Hyvönen M, Morgavi DP, Janssen DB, Hollfelder F. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nat Commun. 2015 Dec 7;6:10008. doi: 10.1038/ncomms10008. PMID: 26639611; PMCID: PMC4686663.


