Over the past decade, the field of single-cell RNA sequencing (scRNA-seq) has witnessed a remarkable enhancement in its capacity to analyze and decipher the transcriptomes of individual cells. Central to this progress is the emergence and widespread integration of droplet microfluidics technology, which has increased the throughput of single cell sequencing campaigns by more than tenfold. Despite this profound advancement, certain challenges inherent to high-throughput droplet microfluidics techniques have become evident. For example, as the scale of cell processing expands, the potential for introducing background noise into the data becomes amplified. Samples that are complex due to damaged cells, debris, or contaminants are particularly susceptible to confounding artefacts that hinder accurate data analysis. Moreover, single-cell encapsulation within droplets is dictated by the Poisson distribution, sometimes leading to the presence of cell multiplets, which can compromise data integrity. Here, researchers from the University of Cambridge present a novel workflow to address these challenges, called spinDrop (sorting and picoinjection inDrop). This workflow isolates droplets containing single viable cells, intact nuclei, or specific cell types prior to scRNA-seq, leading to a significant reduction in background noise. It also addresses limitations in the sensitivity of existing droplet-based scRNA-seq workflows, such as inDrop and Drop-seq, by decoupling cell lysis and reverse transcription steps, reporting here a 5x increase in gene detection rates.
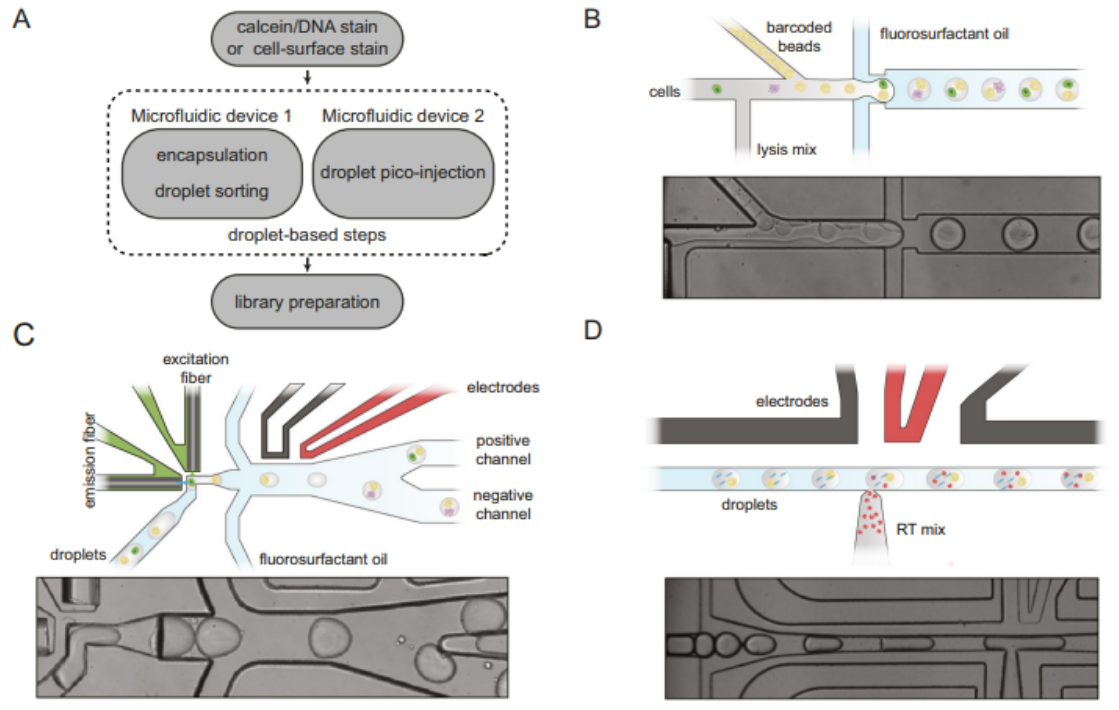
The goal of the spinDrop workflow is to maximize the information gleaned from high-throughput scRNA-seq experiments by reducing data artefacts and improving RNA capture efficiency. To this end, the workflow makes use of two microfluidic devices. The first device employs fluorescence-activated droplet sorting (FADS) to select for, sort, and collect droplets containing single viable cells while excluding empty droplets or those with debris and damaged cells. Unlike using FACS or MACS to pre-sort live cells, in-line FADS acts directly during the encapsulation stage, ensuring cell viability at this critical moment. And, in contrast to using post-processing steps or computational corrections to deal with data artefacts, in-line FADS ensures that only droplets harboring targeted, viable single cells undergo sequencing, offering a more efficient use of sequencing resources. FADS accommodates various enrichment criteria based on the chosen detection method; for example, staining with a viability dye like Calcein-acetoxymethyl (AM) can be used to select for live cells, staining with a DNA dye such as Vybrant Green can detect intact nuceli or be used for fixed cell sorting, and fluorescently labeled antibodies against cell surface markers can be used to select for specific cell types.
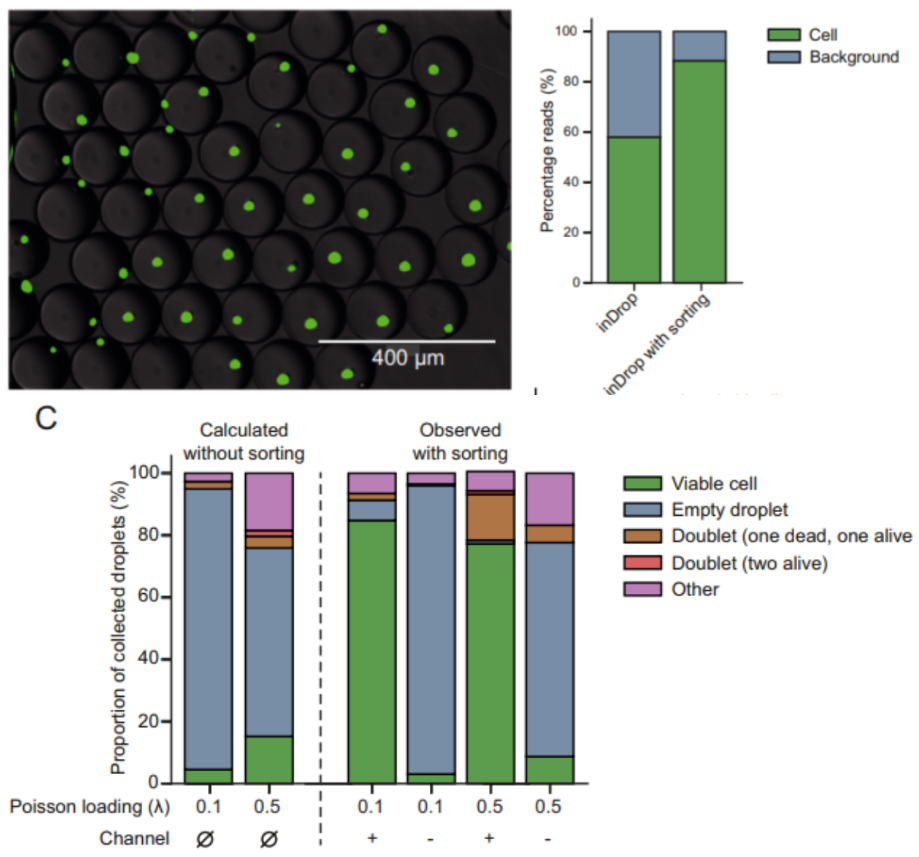
In validating the efficacy of FADS for enriching single viable cells, HEK293T cells were stained with Calcein-AM, then encapsulated in droplets with lysis reagents and polyacrylamide microgels containing inDrop v3 cellular barcodes. These droplets were then immediately sorted based on fluorescence; any droplets containing a viable cell would emit green fluorescence when excited by blue light and trigger the sorting mechanism, while damaged cells or empty droplets would not emit fluorescence and flow into the waste channel. Upon imaging, it was found that 96.1% of the positive droplets contained single viable cells, vastly outperforming the expected values without sorting (9.0% at λ = 0.1). To represent a challenging sample with many damaged cells, a 1:1 mixture of dead and alive HEK293T was used during encapsulation. Sorting this sample led to a 19-fold enrichment of single viable cells (84.8% of droplet population). To test the ability of FADS to discard droplets containing cell multiplets, this procedure was repeated at a higher Poisson loading (λ = 0.5), and an upper fluorescence threshold was set to exclude brighter droplets with multiple cells. This resulted in 77.2% single viable cells in the sorted droplets. The study further assessed the capacity of FADS to enrich specific cell types by using fluorescently labeled antibodies (IGM, CD19, and CD45R) to label a population of mouse peripheral blood mononuclear cells. FADS resulted in a 1.8x enrichment in B cells and the complete depletion of monocytes, which accounted for 9% of the droplet population without sorting. When sequenced, samples that were sorted had 88.4% of reads match a cell barcode, compared to 58.0% for samples that were not sorted. Thus, sorting droplets prior to scRNA-seq can reduce background noise and enable more deterministic sequencing of single, viable cells of research interest. It also ensures a more efficient utilization of sequencing resources by discarding empty droplets or droplets containing cell debris, damaged cells, and undesired cell types prior to the sequencing run.

Current droplet-based scRNA-seq workflows do not change the droplet contents following encapsulation. As a result, certain lysis conditions, such as proteinase K and heat denaturation, cannot be used, as these methods are incompatible with enzymatic reverse transcription. Incorporating a picoinjection module into the spinDrop protocol resolves this issue, as reverse transcriptase can be added after cell lysis is complete. This improvement allows for more effective lysis conditions that will increase RNA yields and improve sensitivity. So, following FADS, the single viable cells within droplets undergo lysis and a heat treatment. The droplets containing single-cell lysate are then loaded into a microfluidic device with picoinjection capabilities, which adds the reverse transcription mix into each droplet. Separating the cell lysis and reverse transcription steps using this multi-step workflow resulted in a fivefold increase in gene detection rates compared to the conventional inDrop workflow, bringing the sensitivity of spinDrop on par with that of the 10x Chromium v2 formulation. The increased sensitivity of the spinDrop workflow is paired with a significant cost advantage, reducing the overall sequencing cost per cell by 60%. The integration of improved RNA capture efficiency, reduced background noise, and cost-effectiveness positions spinDrop as a promising platform for large-scale single-cell profiling endeavors. Moreover, this workflow is well-poised to complement microfluidic techniques that extend beyond transcriptome sequencing, such as ATAC-seq or multi-omic profiling.


