Red Sea microbial life is subjected to extremely challenging environmental conditions, including high salinity, high temperature and pressure, and low nutritional resource availability. As a result, the microbial proteins that have evolved under these selective pressures are likely to harbor new and unique functional activities that may prove valuable in industrial applications. Yet, culturing microorganisms directly from the environment is challenging, which has limited the ability to explore the metabolic potential of these organisms. Functional metagenomics is an approach that bypasses the need for culturing microorganisms and instead involves extracting DNA directly from environmental samples and constructing expression libraries that represent the genetic material of the microbial community. Due to the massive size of these metagenomic libraries, high-throughput screening capabilities are of paramount importance to the successful identification of novel genes or enzymes with industrial applications. In this study, Alma’abadi et al use a high-throughput droplet microfluidics platform to screen a large metagenomic library for genes encoding novel lipolytic enzymes, which are in high demand for industrial applications such as biodiesel generation and the biodegradation of plastics.
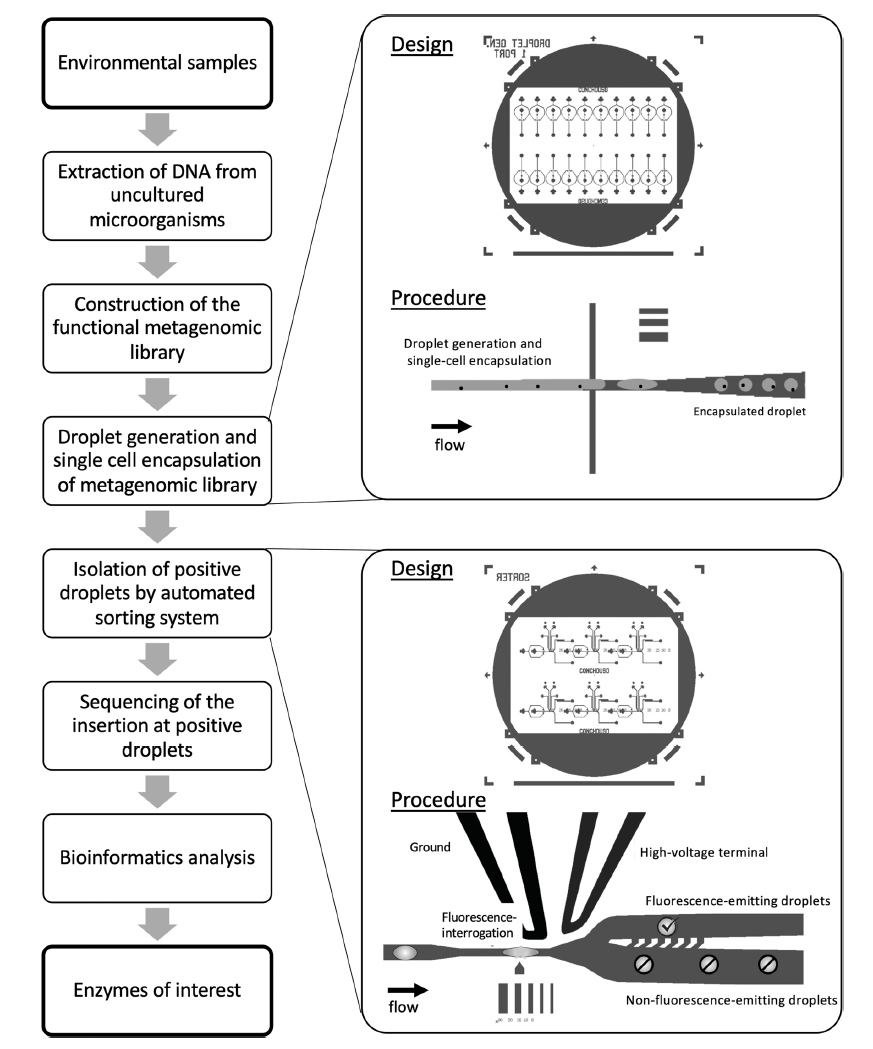
To construct the functional metagenomic library, DNA was extracted from Red Sea surface water samples, subjected to gel electrophoresis, and inserted into fosmids. The resulting library contained 85,000 clones representing the Red Sea microbial community. Individual cells were encapsulated in droplets with standard LB growth media and lipase-specific substrates tagged with fluorescent markers. The individual cells were allowed to incubate in droplets at 37°C for 24 hours, which provided the cells with sufficient time to produce lipolytic enzymes and for those enzymes to interact with the fluorogenic substrate. After the incubation period, the droplets were inserted into the droplet sorter for high-throughput fluorescence-activated droplet sorting (FADS). If a droplet contained enzymes with lipolytic activity, the fluorogenic substrate would be cleaved and fluorescence would be emitted. These droplets could then be automatically selected via dielectrophoresis in the droplet sorter. E.coli containing a plasmid expressing the lipase gene Lip32Nc was used as a positive control.
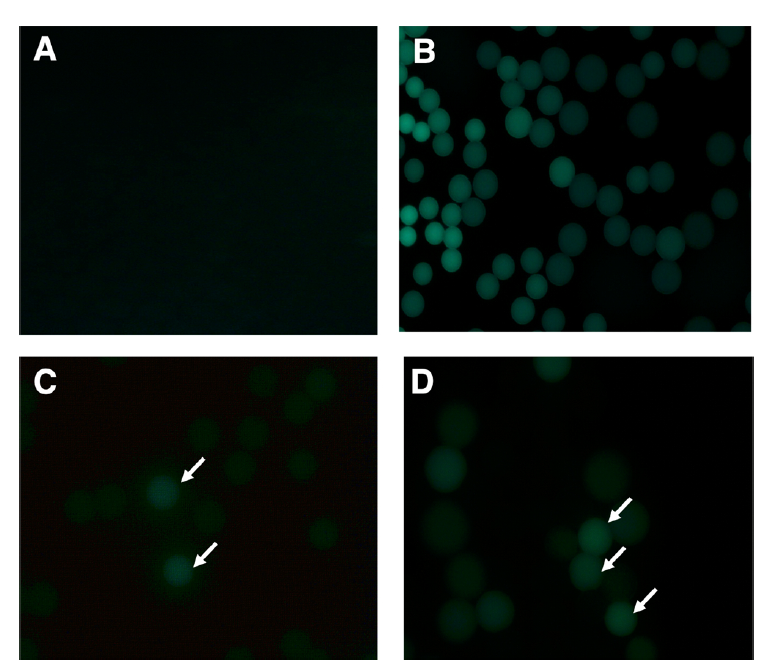
Of the >1,000,000 droplets inserted into the droplet sorter for FADS, 27 positive droplets were selected that contained enzymes with lipolytic activities. This demonstrates the value of using a high-throughput approach when screening metagenomic libraries for novel functions, as positive hits are often quite rare. Metagenomic fragments from 25 samples were sequenced using whole-genome shotgun sequencing, assembled, and annotated. Three potential lipase genes were identified using a BLASTp homology search (LP001, LP002, LP003). LP001 is a putative thermostable monoacylglycerol lipase with a sequence most like Kocuria sp. LP002 is likely a monoacylglycerol lipase, also with sequence homology to a gene from Kocuria sp. LP003 is a putative lipase with lipid degradation activity. Due to its thermostability, LP001 is the most likely candidate for industrial applications. The authors noted that the remaining 22 selected droplets still produced fluorescence, which indicates lipolytic activity. These droplets may thus contain novel genes with potentially valuable features.

The lipolytic activity of the 25 droplets was confirmed using the Spirit Blue Agar assay. Bacteria that produce lipase will hydrolyze the triglycerides in the agar surrounding them, resulting in a visible clear zone surrounding the colony. Each of the 25 droplets, including the 22 without known lipolytic enzymes, showed lipase activity in this assay. This observation further supports the hypothesis that these droplets contain new lipase genes, or novel lipolytic functions of known genes. Further investigation of these unknown genes is necessary to understand their functional characteristics and potential applications.
- Alma'abadi A, Behzad H, Alarawi M, Conchouso D, Saito Y, Hosokawa M, Nishikawa Y, Kogawa M, Takeyama H, Mineta K, Gojobori T. Identification of lipolytic enzymes using high-throughput single-cell screening and sorting of a metagenomic library. N Biotechnol. 2022 Sep 25;70:102-108. doi: 10.1016/j.nbt.2022.05.006. Epub 2022 May 28. PMID: 35636700.


