The production of synthetic DNA molecules, such as single-stranded DNA (ssDNA) oligonucleotides, is essential to applications in molecular biology and bioengineering, for instance, as donor DNA for CRISPR-Cas9 genome editing, as scaffolding for DNA nanotechnology, as a medium for archival data storage, and as a drug delivery vehicle capable of distributing molecular payloads to cells. Currently, ssDNA can be produced chemically using phosphoramidite synthesis, which is efficient for short oligonucleotides but suffers from poor yields as oligonucleotide length increases. Enzymatic synthesis methods, such as catalysis by terminal deoxynucleotidyl transferase (TdT) and asymmetric PCR (aPCR), can be used to produce longer ssDNA oligonucleotides but pose challenges that limit their current applications, namely, the need to control the addition of single bases in TdT synthesis and the extensive experimentation required to optimize aPCR protocols. In this paper, the authors suggest an improvement to enzymatic synthesis of longer ssDNA oligonucleotides using in vitro transcription (IVT). Using directed evolution assisted by fluorescence-activated droplet sorting, they selected for a T7 RNA polymerase (T7 RNAP) with reduced substrate specificity that is capable of directly incorporating DNA, rather than RNA, during the IVT reaction, which would enable the production of DNA oligonucleotides without a reverse transcription step. Here, the authors report that the identified T7 RNAP mutants produce high-quality chimeric ssDNA oligonucleotides quickly and efficiently, and that these ssDNA oligonucleotides can serve as PCR primers.
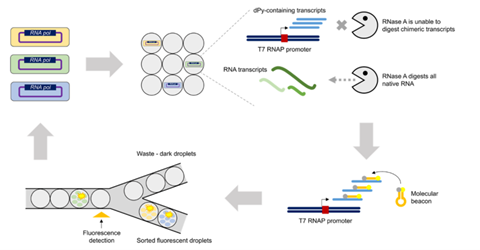
Seventy-seven amino acid positions within the substrate binding pocket of T7 RNAP were subjected to site-saturation mutagenesis, resulting in a mutant library consisting of 1463 unique variants. These variants were cloned into expression vectors and transformed into E. coli cells; the presence of the desired mutations was verified with Sanger sequencing. The T7 RNAP variant Y639F, which has been previously reported to incorporate deoxyribopyrimidine triphosphates (dPy) and thus would be expected to show enrichment in the designed in vitro selection scheme, was also cloned into expression vectors and transformed into E. coli cells as a positive control. Individual E. coli cells were then coencapsulated with lysozyme, IVT reagents and fluorescent molecular beacon tags into water-in-oil droplets using the ONYX droplet generator. To ensure selection for T7 RNAP variants with reduced substrate specificity capable of incorporating dPYs, dPYs replaced ribopyrimidines in the IVT reagents. In addition, RNase A was added to digest native RNA transcripts, leaving only dPY-containing transcripts. Then, the molecular beacons were hybridized so that target IVT products could be identified and sorted via fluorescence using the STYX droplet sorter.
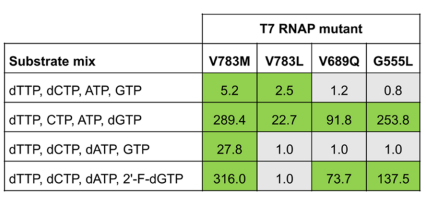
After two rounds of selection, the authors observed enrichment of the internal control Y639F mutation as well as enrichment of newly identified mutants V783M (54x enriched), V783L (9x enriched), V689Q (12x enriched), and G555L (8x enriched). These T7 RNAPs represented variants capable of producing chimeric DNA oligonucleotides during the IVT reaction. These mutants were tested for their ability to synthesize transcripts containing deoxynucleotide triphosphates (dTTP, dCTP, dATP, and 2’-fluoro-dGTP). Compared to the amount of transcript produced by wild type T7 RNAP under the same conditions, V783M mutant produced the highest amount of full-length target transcripts in all conditions, with a yield ratio of 316.0 when dTTP, dCTP, dATP, and 2’-F-dGTP were used as substrates for IVT. Mutants G555L (yield ratio 137.5) and V689Q (yield ratio 73.7) also showed acceptable performance with these substrates. Comparison of the V7833M mutant to internal control Y639F revealed similar activity and efficiency between these two T7 RNAPs. Moreover, the activity of double mutants V783M/V689Q and V783M/G555L was three times higher than single V783M or Y639F mutants.
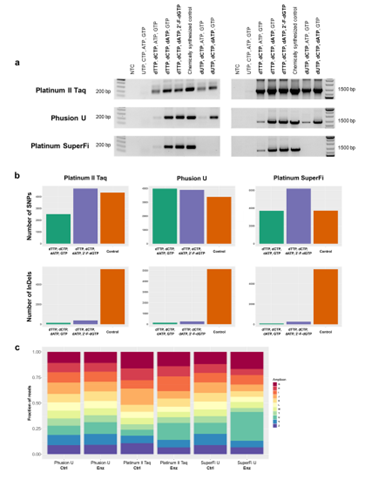
Proof-of-principle experiments were conducted to assess the viability of the IVT-generated ssDNA oligonucleotides for use as PCR primers in single-plex and multiplex PCR. Mutant V783M was used to synthesize 42 nt oligonucleotides with various combinations of ribonucleotides and deoxyribonucleotides to be used as a forward primer in single-plex PCR with either 0.2 kb or 1.4 kb target amplicon. Taq DNA polymerase was able to use IVT-synthesized oligonucleotides made with any combination of ribo- and deoxyribonucleotides other than pure RNA transcript, and proofreading polymerases produced amplicons when the IVT-synthesized oligonucleotides contained at least three deoxyribonucleotides. The resulting 0.2 kb amplicons were sequenced to assess mutational profiles within the locus covered by the IVT-produced primers; compared with chemically synthesized controls, there was no difference in the number of single-nucleotide polymorphisms seen in the IVT-synthesized primers and there were significantly fewer insertions and deletions, confirming the fidelity of the IVT-directed oligonucleotide synthesis using the V783M mutant T7 RNAP. The authors also demonstrated the feasibility of synthesizing 11 different oligonucleotide primers in a single IVT reaction that could then be used for multiplex PCR with a careful selection of PCR conditions. Thus, the evolved T7 RNAPs could be used for fast, efficient, and inexpensive one-pot synthesis of multiple DNA oligonucleotides in parallel. Combined with the ability to produce longer and more complex oligonucleotides, the evolved T7 RNAPs represent a significant improvement in enzymatic synthesis of ssDNA.



