Droplet microfluidic technology is an innovative approach that has revolutionized many areas of research, including microbial analysis. By compartmentalizing individual microbes into droplets, this approach enables researchers to perform highly sensitive and specific identification of bacteria that make up the human and environmental microbiomes. Single-cell microbial research enhances our understanding of microbial diversity and interactions, with the potential to uncover new insights into the ecological and evolutionary dynamics of microbial communities. With its ability to provide strain-level resolution, droplet microfluidics provides a high-throughput, cost-effective, and accurate analysis of individual microbial genomes.
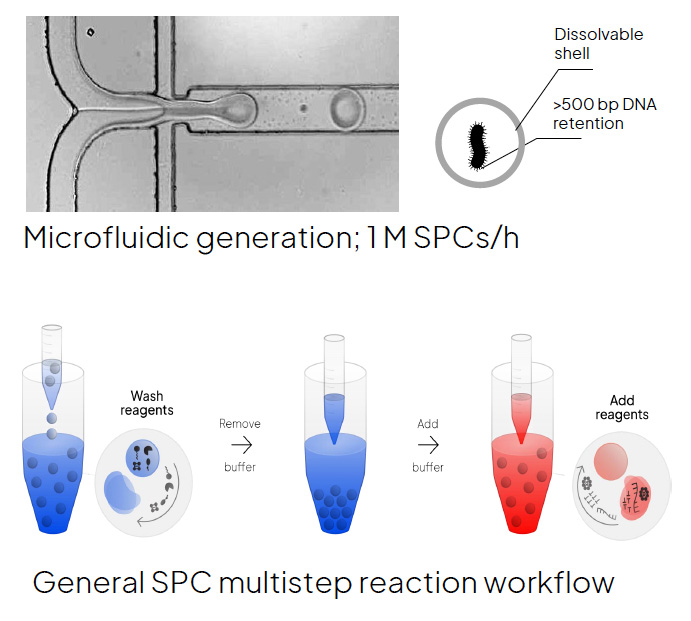
However, analyzing bacteria and other microorganisms can pose many challenges for classic droplet microfluidic techniques. For example, the harsh cell lysis required to access bacterial genomes not only interferes with droplet stability but is also often incompatible with downstream molecular biology reactions. Semi-permeable capsules (SPCs) are a breakthrough technology designed to support high-throughput multi-step molecular analysis, even those that require a harsh lysis step, without loss of single-cell compartmentalization. Semi-Permeable Capsules (SPCs) have an aqueous core that is surrounded by a hydrogel shell that functions as a size-selective membrane, allowing the exchange of small reaction components such as enzymes, primers, and salts for bacterial lysis and amplification of single bacterial genomes.
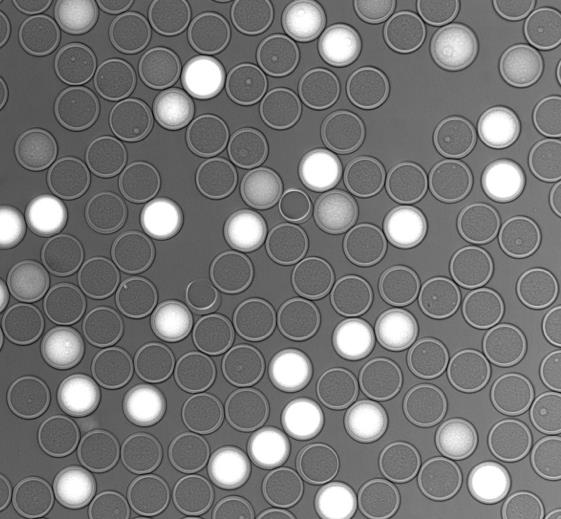
Each SPC can be considered an individual reaction vessel that can be easily manipulated and monitored, allowing for highly efficient amplification reactions. Once generated, the SPCs are stable under any reaction conditions and are suitable for any downstream reaction, which can be performed in a bulk-like manner on millions of SPCs in parallel using standard laboratory equipment. Here, single-amplified genomes (SAGs) in SPCs that have captured a bacterium were visualized with SYTO9 green fluorescent nucleic acid stain. SAGs can then be barcoded and prepped into libraries prior to sequencing using next-generation sequencing (NGS) platforms. SPC technology enables the barcoding and sequencing of 1,000 to 100,000 individual bacterial cells in a single experiment, permitting the high-throughput strain-level identification of microbes.
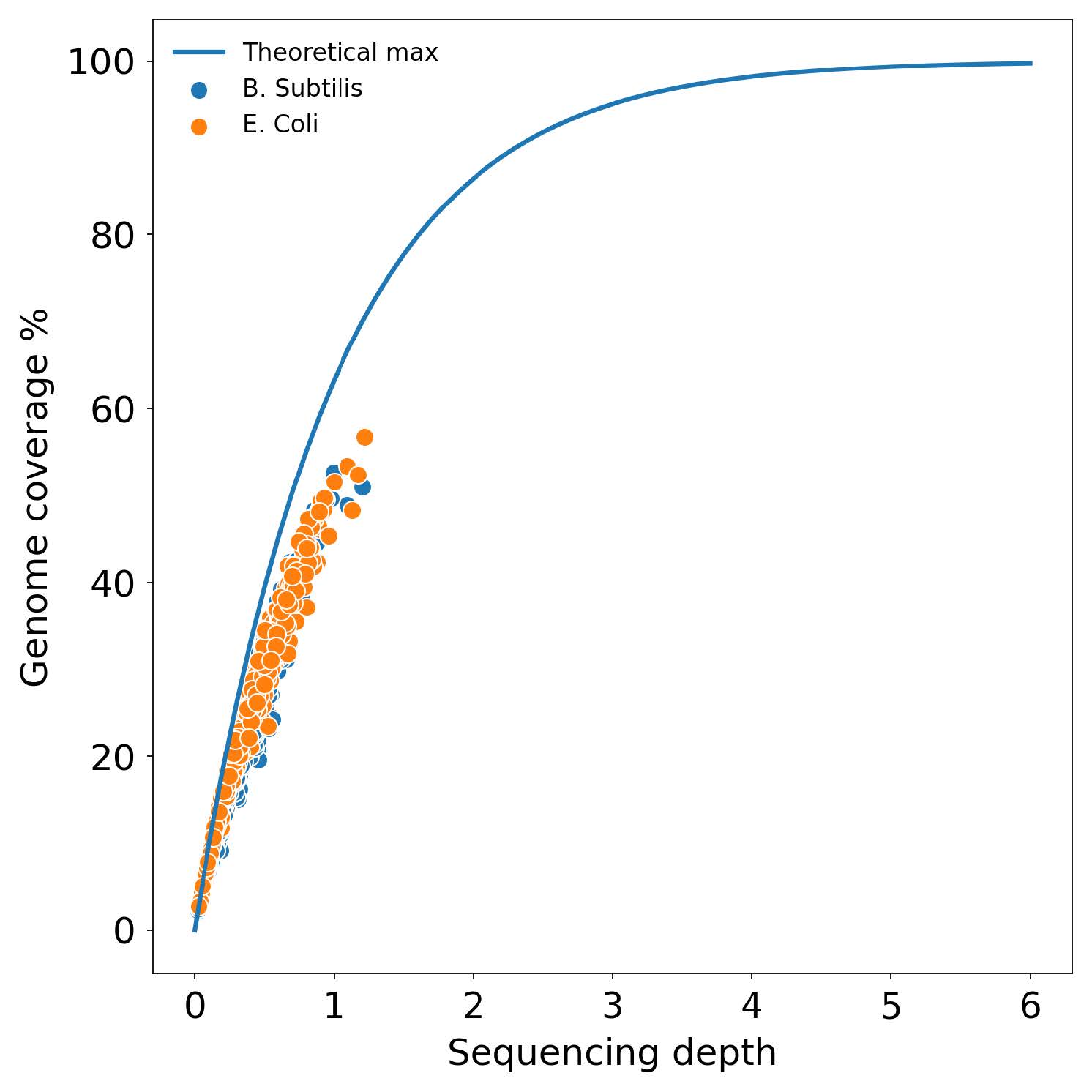
During an experiment that employed SPC technology to screen 1638 genomes of Escherichia coli and Bacillus subtilis, only 0.24% of the single-amplified genomes (SAGs) were found to have low purity. This was determined by excluding genomes that had <99% reads mapped to either bacterium. Depending on the sequencing depth, one SAG can result in up to 60% genome coverage. This suggests that multiple SAGs may be combined to achieve 100% genome coverage. SPC technology is an ideal platform for multi-step molecular analysis, including microbial single-cell sequencing, and has been shown to enable the barcoding and sequencing of up to a hundred thousand individual microbes with low contamination rates. This technology supports the high-throughput sequencing of microbes, including unculturable bacteria, allowing researchers to investigate microbial community structure and functional capacity at the strain level.