The COVID-19 pandemic caused by SARS-CoV-2 has resulted in a devastating global health crisis, with millions of infections and deaths reported. To combat the virus, mRNA-based vaccines developed by Moderna and Pfizer-BioNTech were swiftly deployed worldwide. These vaccines demonstrated efficacy in phase-III trials by inducing robust immune responses that effectively prevent severe COVID-19 and mortality. However, subsequent studies conducted after vaccine distribution have observed a significant decline in neutralizing antibody titers in the months following vaccination. Single-cell studies of antibody-secreting cells (ASCs) can thus offer valuable insights for understanding the duration of immunity and for optimizing vaccination protocols. Here, researchers from The Pasteur Institute report a droplet microfluidics-based kinetic analysis of single ASCs in cohorts of BNT162b2 (Pfizer-BioNTech)-vaccinated individuals. The single-cell bioassay, called DropMap, provides a qualitative assessment of the humoral immune response following vaccination, including estimations of the secretion rate, specificity, and affinity for SARS-CoV-2 receptor-binding domains of human IgG secreted by circulating ASCs (I.e., plasmablasts and plasma cells). As droplet microfluidics platforms offer massive parallelization capabilities, the use of this assay can reduce the workload, time, and costs associated with single-cell immune monitoring, thereby enhancing the efficiency and scalability of these analyses.
This study followed two cohorts of individuals following vaccination with the BNT162b2 mRNA vaccine. The first cohort (COVID-recovered) contained 18 patients who received one vaccine dose following recovery from COVID-19 infection (mean 309 ± 44.6 days post-infection) in accordance with French guidelines. The second cohort (COVID-naive) contained 11 healthcare workers with no clinical history of COVID-19 and no serological evidence of previous SARS-CoV-2 infection. These individuals received a primary vaccination and a booster vaccination (mean 27.7 ± 1.8 days apart). Samples were collected from both cohorts prior to vaccination, shortly after the final vaccine dose was received, and 2 months after the final dose was received. Peripheral blood mononuclear cells were isolated from the samples and suspended at a cell density to achieve less than or equal to one cell per droplet. Cells were co-encapsulated with paramagnetic nanoparticles and fluorescently labeled reporter proteins via a co-flow droplet generator. Droplets were then injected into an observation chamber for the single-cell DropMap assay.
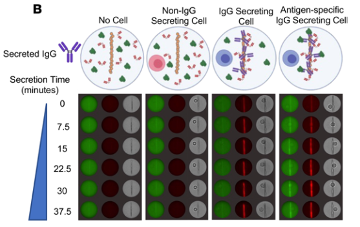
The DropMap assay is centered around assessing fluorescence relocation to the beadline, a clearly distinguishable pattern formed within each droplet by the paramagnetic beads when a magnetic field is applied to the observation chamber. The paramagnetic beads are coated with an IgG capture reagent, thus acting as a physical surface for the fluorescence relocation assay. If the beads have bound antibodies, the red fluorescence (AlexaFluor647-labeled fragment of rabbit anti-human IgG) will relocate to the beadline; if the bound antibodies are specific to the SARS-CoV-2 receptor binding domain (RBD), the green fluorescence (fluorophore-conjugated RBD) will relocate to the beadline. For each time point during the observation period (every 7.5 min for 37.5 min), the authors measured the fluorescence signals from the beadline and the entire droplet without the beadline. They compared the fluorescence signals to calibration curves, which were generated using known concentrations of monoclonal anti-RBD IgG with known affinity. By comparing the signals to the calibration curves, they could estimate the rate at which IgG antibodies were being secreted and how well these antibodies were binding to the SARS-CoV-2 RBD.< /p>

Between 10,000-20,000 single PMBCs were analyzed from each patient sample. Previous COVID-19 infection (>6 months ago) did not result in any significant difference in the number of high-affinity IgG-secreting cells (IgG-SC) circulating in the blood prior to vaccination. Following vaccination with the BNT162b2 mRNA vaccine, both cohorts experienced an expansion and contraction of RBD-specific IgG-SC, with a majority (>65%) of the RBD-specific plasmablast pool being lower affinity and with high-affinity plasmablasts disappearing quickly. COVID-recovered individuals had a higher IgG secretion rate prior to vaccination than COVID-naive individuals. While this secretion rate remained relatively unchanged following vaccination for COVID-recovered individuals, COVID-naive individuals experienced a significant increase in IgG secretion rate >6 weeks after vaccination (p < 0.05), to a rate that matched that of the COVID-recovered cohort. These insights into the prevalence and quality of circulating IgG-SC may be combined with data from other single-cell technologies, such as B cell-receptor sequencing, single-cell transcriptomics, and LC-MS/MS proteomics, to inform the optimization of vaccine designs and vaccination protocols.


